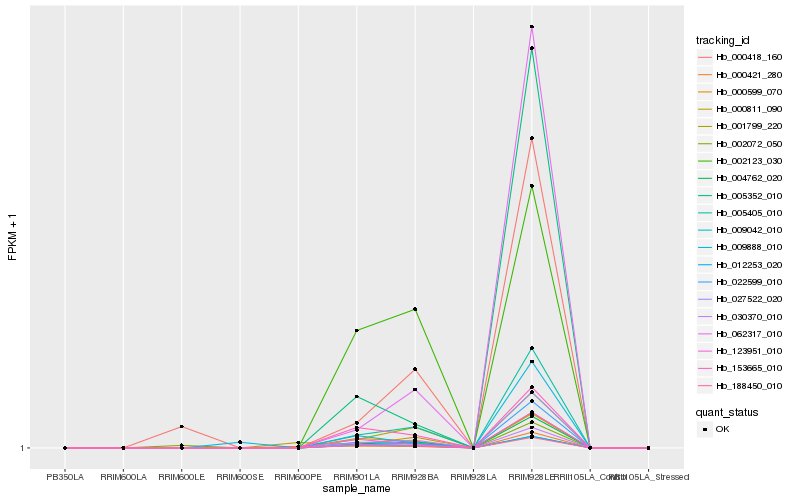
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004762_020 |
0.0 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 2 |
Hb_005405_010 |
0.0094682534 |
- |
- |
- |
| 3 |
Hb_188450_010 |
0.0934191584 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 4 |
Hb_009042_010 |
0.0943010227 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 5 |
Hb_000599_070 |
0.1139888214 |
- |
- |
PREDICTED: patellin-4-like [Jatropha curcas] |
| 6 |
Hb_022599_010 |
0.1140878052 |
- |
- |
Uncharacterized protein TCM_010507 [Theobroma cacao] |
| 7 |
Hb_030370_010 |
0.1373882126 |
- |
- |
PREDICTED: uncharacterized protein LOC104896222 [Beta vulgaris subsp. vulgaris] |
| 8 |
Hb_123951_010 |
0.1377108231 |
- |
- |
PREDICTED: uncharacterized protein LOC104894399 [Beta vulgaris subsp. vulgaris] |
| 9 |
Hb_153665_010 |
0.1433660706 |
- |
- |
PREDICTED: uncharacterized protein LOC104245945 [Nicotiana sylvestris] |
| 10 |
Hb_062317_010 |
0.1458094495 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_005352_010 |
0.1614181001 |
- |
- |
- |
| 12 |
Hb_000418_160 |
0.1615433234 |
- |
- |
- |
| 13 |
Hb_027522_020 |
0.1715868091 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 14 |
Hb_001799_220 |
0.1983645677 |
transcription factor |
TF Family: AP2 |
Ethylene-responsive transcription factor, putative [Ricinus communis] |
| 15 |
Hb_000811_090 |
0.2054699853 |
- |
- |
hypothetical protein VITISV_043745 [Vitis vinifera] |
| 16 |
Hb_002123_030 |
0.2086616113 |
- |
- |
hypothetical protein VITISV_003767 [Vitis vinifera] |
| 17 |
Hb_000421_280 |
0.2127518064 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 18 |
Hb_002072_050 |
0.2141136264 |
- |
- |
PREDICTED: uncharacterized protein LOC104894399 [Beta vulgaris subsp. vulgaris] |
| 19 |
Hb_009888_010 |
0.2142146233 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 20 |
Hb_012253_020 |
0.2155549592 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |