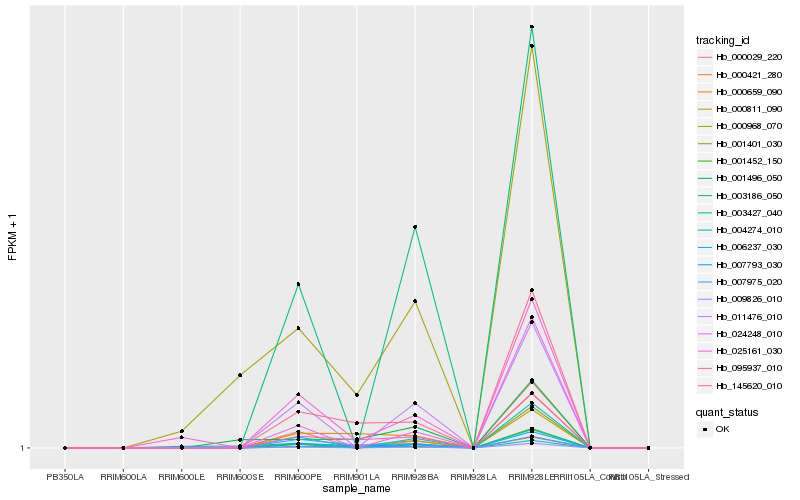
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_095937_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 2 |
Hb_145620_010 |
0.0422387721 |
- |
- |
hypothetical protein VITISV_019786 [Vitis vinifera] |
| 3 |
Hb_006237_030 |
0.0738036338 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 4 |
Hb_000659_090 |
0.0775328936 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 5 |
Hb_000421_280 |
0.1253418468 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 6 |
Hb_000811_090 |
0.1280372795 |
- |
- |
hypothetical protein VITISV_043745 [Vitis vinifera] |
| 7 |
Hb_025161_030 |
0.1971689003 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001496_050 |
0.2142356475 |
- |
- |
yth domain-containing protein, putative [Ricinus communis] |
| 9 |
Hb_007975_020 |
0.2156916417 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 10 |
Hb_001452_150 |
0.2180479227 |
- |
- |
PREDICTED: uncharacterized protein LOC104879858 [Vitis vinifera] |
| 11 |
Hb_003427_040 |
0.2182229553 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 12 |
Hb_009826_010 |
0.2184027166 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 13 |
Hb_004274_010 |
0.2189294171 |
- |
- |
PREDICTED: uncharacterized protein LOC104451199 [Eucalyptus grandis] |
| 14 |
Hb_000968_070 |
0.2225959665 |
- |
- |
- |
| 15 |
Hb_024248_010 |
0.2326244457 |
- |
- |
PREDICTED: uncharacterized protein LOC105779006 [Gossypium raimondii] |
| 16 |
Hb_000029_220 |
0.2359055156 |
- |
- |
PREDICTED: pre-mRNA-splicing factor SLU7-A-like [Jatropha curcas] |
| 17 |
Hb_011476_010 |
0.2383275545 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 18 |
Hb_001401_030 |
0.2475913427 |
- |
- |
PREDICTED: uncharacterized protein LOC105649622 [Jatropha curcas] |
| 19 |
Hb_003186_050 |
0.2476469393 |
- |
- |
- |
| 20 |
Hb_007793_030 |
0.2500868567 |
- |
- |
OSJNBa0074B10.6 [Oryza sativa Japonica Group] |