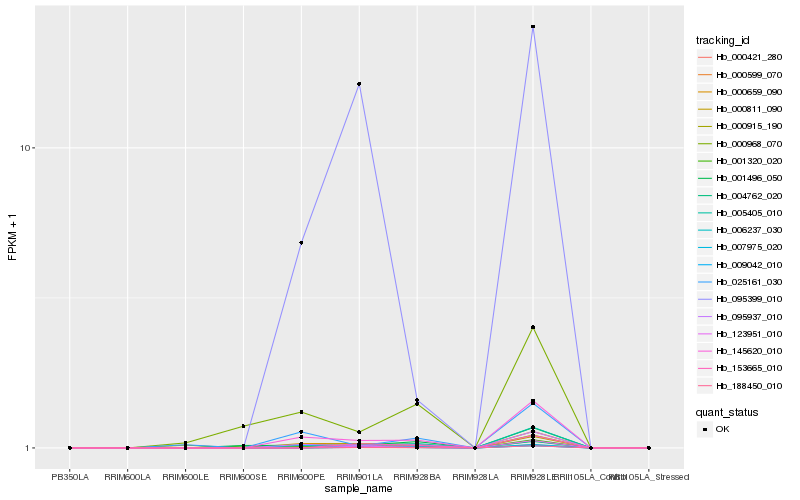
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006237_030 |
0.0 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 2 |
Hb_000659_090 |
0.0415855642 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 3 |
Hb_095937_010 |
0.0738036338 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 4 |
Hb_145620_010 |
0.0824303991 |
- |
- |
hypothetical protein VITISV_019786 [Vitis vinifera] |
| 5 |
Hb_000421_280 |
0.1365803925 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 6 |
Hb_000811_090 |
0.178129734 |
- |
- |
hypothetical protein VITISV_043745 [Vitis vinifera] |
| 7 |
Hb_007975_020 |
0.2305910487 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 8 |
Hb_001496_050 |
0.2309451882 |
- |
- |
yth domain-containing protein, putative [Ricinus communis] |
| 9 |
Hb_000915_190 |
0.2392367227 |
- |
- |
- |
| 10 |
Hb_025161_030 |
0.2479681447 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 11 |
Hb_188450_010 |
0.2494065334 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 12 |
Hb_009042_010 |
0.2494067015 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 13 |
Hb_000599_070 |
0.2502307993 |
- |
- |
PREDICTED: patellin-4-like [Jatropha curcas] |
| 14 |
Hb_000968_070 |
0.2516444765 |
- |
- |
- |
| 15 |
Hb_153665_010 |
0.2542600424 |
- |
- |
PREDICTED: uncharacterized protein LOC104245945 [Nicotiana sylvestris] |
| 16 |
Hb_001320_020 |
0.2622373306 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103696607 [Phoenix dactylifera] |
| 17 |
Hb_123951_010 |
0.2627430075 |
- |
- |
PREDICTED: uncharacterized protein LOC104894399 [Beta vulgaris subsp. vulgaris] |
| 18 |
Hb_095399_010 |
0.2651700768 |
- |
- |
cytochrome b/f complex subunit IV [Sphagnum fimbriatum] |
| 19 |
Hb_004762_020 |
0.265500762 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 20 |
Hb_005405_010 |
0.2668744862 |
- |
- |
- |