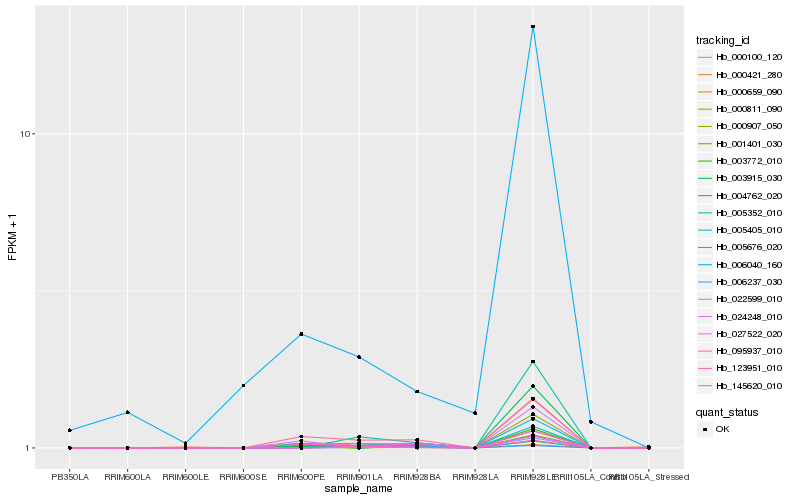
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000421_280 |
0.0 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 2 |
Hb_145620_010 |
0.1008035176 |
- |
- |
hypothetical protein VITISV_019786 [Vitis vinifera] |
| 3 |
Hb_000811_090 |
0.1201906121 |
- |
- |
hypothetical protein VITISV_043745 [Vitis vinifera] |
| 4 |
Hb_095937_010 |
0.1253418468 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 5 |
Hb_006237_030 |
0.1365803925 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 6 |
Hb_000659_090 |
0.1687178683 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 7 |
Hb_003915_030 |
0.1913086847 |
- |
- |
PREDICTED: uncharacterized protein LOC104901272 [Beta vulgaris subsp. vulgaris] |
| 8 |
Hb_022599_010 |
0.2061825437 |
- |
- |
Uncharacterized protein TCM_010507 [Theobroma cacao] |
| 9 |
Hb_005352_010 |
0.206414988 |
- |
- |
- |
| 10 |
Hb_004762_020 |
0.2127518064 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 11 |
Hb_001401_030 |
0.2159688901 |
- |
- |
PREDICTED: uncharacterized protein LOC105649622 [Jatropha curcas] |
| 12 |
Hb_027522_020 |
0.2164309344 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 13 |
Hb_005405_010 |
0.2165552549 |
- |
- |
- |
| 14 |
Hb_123951_010 |
0.2174767398 |
- |
- |
PREDICTED: uncharacterized protein LOC104894399 [Beta vulgaris subsp. vulgaris] |
| 15 |
Hb_000907_050 |
0.2220003726 |
- |
- |
- |
| 16 |
Hb_024248_010 |
0.2233361089 |
- |
- |
PREDICTED: uncharacterized protein LOC105779006 [Gossypium raimondii] |
| 17 |
Hb_005676_020 |
0.2245910121 |
transcription factor |
TF Family: ERF |
hypothetical protein RCOM_1314900 [Ricinus communis] |
| 18 |
Hb_000100_120 |
0.2249859926 |
- |
- |
RNA-directed DNA polymerase (Reverse transcriptase); Chromo; Zinc finger, CCHC-type; Peptidase aspartic, active site; Polynucleotidyl transferase, Ribonuclease H fold [Medicago truncatula] |
| 19 |
Hb_003772_010 |
0.2259735529 |
- |
- |
polyprotein [Oryza australiensis] |
| 20 |
Hb_006040_160 |
0.2269070855 |
- |
- |
ribosomal protein S12 (mitochondrion) [Hevea brasiliensis] |