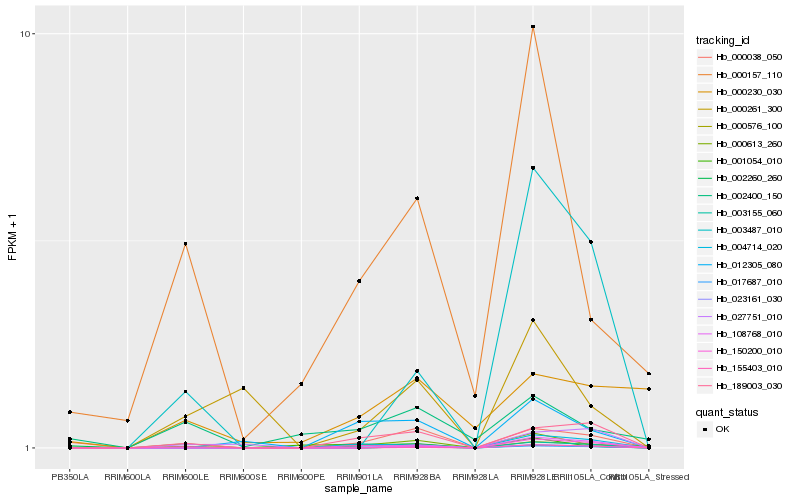
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_189003_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_023161_030 |
0.2259852317 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 3 |
Hb_004714_020 |
0.265325141 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 4 |
Hb_150200_010 |
0.2686828605 |
- |
- |
PREDICTED: uncharacterized protein LOC105632410 [Jatropha curcas] |
| 5 |
Hb_000038_050 |
0.3151690878 |
- |
- |
- |
| 6 |
Hb_012305_080 |
0.3208157924 |
- |
- |
- |
| 7 |
Hb_017687_010 |
0.3436574691 |
- |
- |
retrotransposon protein, putative, Ty3-gypsy subclass [Oryza sativa Japonica Group] |
| 8 |
Hb_000613_260 |
0.3464427888 |
- |
- |
hypothetical protein VITISV_043487 [Vitis vinifera] |
| 9 |
Hb_003487_010 |
0.354987131 |
- |
- |
- |
| 10 |
Hb_155403_010 |
0.3628487082 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 11 |
Hb_027751_010 |
0.3762024414 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 12 |
Hb_108768_010 |
0.3810496563 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |
| 13 |
Hb_000576_100 |
0.3831997119 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 14 |
Hb_003155_060 |
0.3833700421 |
- |
- |
PREDICTED: uncharacterized protein LOC104105697 [Nicotiana tomentosiformis] |
| 15 |
Hb_000230_030 |
0.390733187 |
- |
- |
hypothetical protein B456_013G2021001, partial [Gossypium raimondii] |
| 16 |
Hb_002260_260 |
0.3933347668 |
- |
- |
hypothetical protein VITISV_037667 [Vitis vinifera] |
| 17 |
Hb_000261_300 |
0.3970399002 |
- |
- |
- |
| 18 |
Hb_001054_010 |
0.4051918898 |
- |
- |
hypothetical protein VITISV_011091 [Vitis vinifera] |
| 19 |
Hb_002400_150 |
0.4053534899 |
- |
- |
PREDICTED: uncharacterized protein LOC105650070 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000157_110 |
0.4076938861 |
- |
- |
hypothetical protein Csa_6G439410 [Cucumis sativus] |