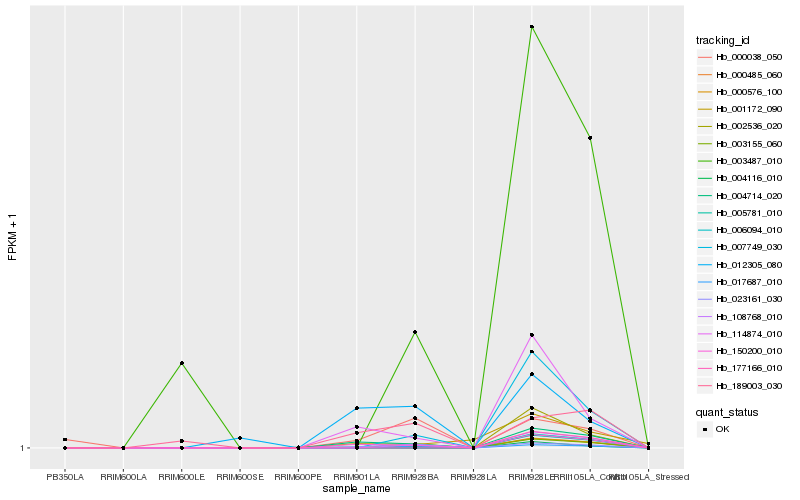
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_150200_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632410 [Jatropha curcas] |
| 2 |
Hb_004714_020 |
0.013006971 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 3 |
Hb_114874_010 |
0.1859744563 |
- |
- |
PREDICTED: uncharacterized protein LOC104226968, partial [Nicotiana sylvestris] |
| 4 |
Hb_023161_030 |
0.216018737 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 5 |
Hb_108768_010 |
0.2604782834 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |
| 6 |
Hb_000576_100 |
0.2604839356 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 7 |
Hb_003155_060 |
0.2604873987 |
- |
- |
PREDICTED: uncharacterized protein LOC104105697 [Nicotiana tomentosiformis] |
| 8 |
Hb_189003_030 |
0.2686828605 |
- |
- |
- |
| 9 |
Hb_012305_080 |
0.2710279785 |
- |
- |
- |
| 10 |
Hb_007749_030 |
0.2826592921 |
- |
- |
- |
| 11 |
Hb_002536_020 |
0.2916714742 |
- |
- |
PREDICTED: uncharacterized protein LOC105801112 [Gossypium raimondii] |
| 12 |
Hb_003487_010 |
0.3065257138 |
- |
- |
- |
| 13 |
Hb_017687_010 |
0.3069032202 |
- |
- |
retrotransposon protein, putative, Ty3-gypsy subclass [Oryza sativa Japonica Group] |
| 14 |
Hb_004116_010 |
0.3155518121 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 15 |
Hb_001172_090 |
0.3218585457 |
- |
- |
PREDICTED: uncharacterized protein LOC104094989 [Nicotiana tomentosiformis] |
| 16 |
Hb_000038_050 |
0.3405788713 |
- |
- |
- |
| 17 |
Hb_006094_010 |
0.3475486272 |
- |
- |
PREDICTED: uncharacterized protein LOC104227769, partial [Nicotiana sylvestris] |
| 18 |
Hb_005781_010 |
0.3475486885 |
- |
- |
PREDICTED: probable terpene synthase 6 [Jatropha curcas] |
| 19 |
Hb_177166_010 |
0.3475529625 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 20 |
Hb_000485_060 |
0.3475540142 |
- |
- |
PREDICTED: uncharacterized protein LOC105641351 [Jatropha curcas] |