| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
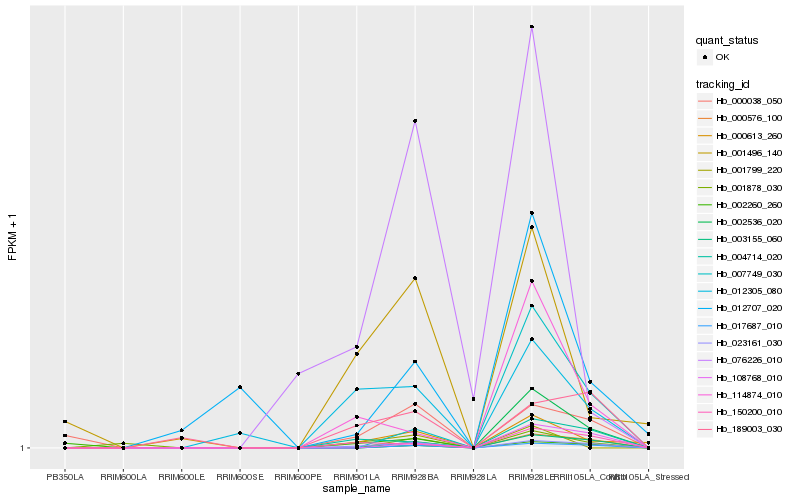
Hb_023161_030 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 2 |
Hb_150200_010 |
0.216018737 |
- |
- |
PREDICTED: uncharacterized protein LOC105632410 [Jatropha curcas] |
| 3 |
Hb_004714_020 |
0.2165885088 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 4 |
Hb_000038_050 |
0.220161457 |
- |
- |
- |
| 5 |
Hb_189003_030 |
0.2259852317 |
- |
- |
- |
| 6 |
Hb_017687_010 |
0.2260945507 |
- |
- |
retrotransposon protein, putative, Ty3-gypsy subclass [Oryza sativa Japonica Group] |
| 7 |
Hb_012305_080 |
0.2304510124 |
- |
- |
- |
| 8 |
Hb_001496_140 |
0.3042298493 |
- |
- |
- |
| 9 |
Hb_108768_010 |
0.3090862109 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |
| 10 |
Hb_000576_100 |
0.3106129645 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 11 |
Hb_003155_060 |
0.3107345313 |
- |
- |
PREDICTED: uncharacterized protein LOC104105697 [Nicotiana tomentosiformis] |
| 12 |
Hb_114874_010 |
0.3217412454 |
- |
- |
PREDICTED: uncharacterized protein LOC104226968, partial [Nicotiana sylvestris] |
| 13 |
Hb_002260_260 |
0.3384162992 |
- |
- |
hypothetical protein VITISV_037667 [Vitis vinifera] |
| 14 |
Hb_076226_010 |
0.3442047604 |
- |
- |
hypothetical protein VITISV_030432 [Vitis vinifera] |
| 15 |
Hb_001878_030 |
0.3460998617 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105126050 [Populus euphratica] |
| 16 |
Hb_007749_030 |
0.3503582251 |
- |
- |
- |
| 17 |
Hb_000613_260 |
0.3518466797 |
- |
- |
hypothetical protein VITISV_043487 [Vitis vinifera] |
| 18 |
Hb_001799_220 |
0.3582680895 |
transcription factor |
TF Family: AP2 |
Ethylene-responsive transcription factor, putative [Ricinus communis] |
| 19 |
Hb_012707_020 |
0.3630951476 |
- |
- |
- |
| 20 |
Hb_002536_020 |
0.3638608984 |
- |
- |
PREDICTED: uncharacterized protein LOC105801112 [Gossypium raimondii] |