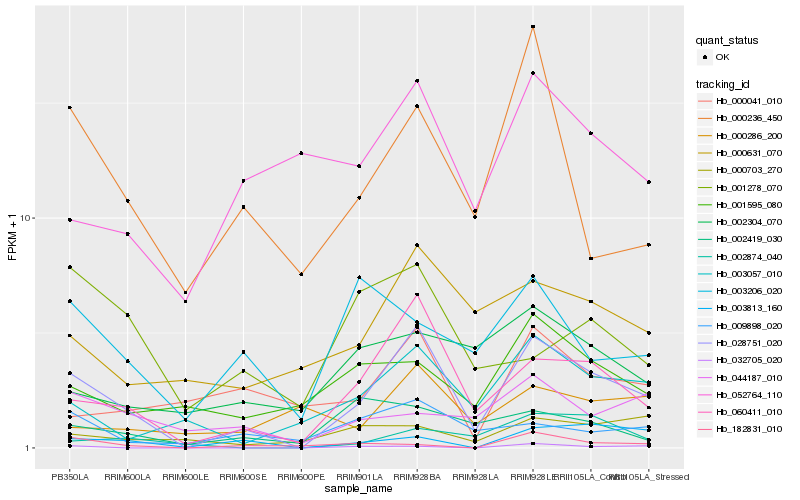
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028751_020 |
0.0 |
- |
- |
PREDICTED: SCY1-like protein 2 [Jatropha curcas] |
| 2 |
Hb_060411_010 |
0.1801276684 |
- |
- |
PREDICTED: uncharacterized protein LOC100246093 [Vitis vinifera] |
| 3 |
Hb_003057_010 |
0.2649295441 |
- |
- |
hypothetical protein JCGZ_06666 [Jatropha curcas] |
| 4 |
Hb_000631_070 |
0.2742795283 |
transcription factor |
TF Family: C2H2 |
PREDICTED: histone deacetylase 5 [Jatropha curcas] |
| 5 |
Hb_009898_020 |
0.2829766411 |
- |
- |
hypothetical protein JCGZ_23951 [Jatropha curcas] |
| 6 |
Hb_000236_450 |
0.2857838197 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 7 |
Hb_003813_160 |
0.2865572585 |
- |
- |
- |
| 8 |
Hb_001595_080 |
0.2910998194 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_052764_110 |
0.298785742 |
- |
- |
PREDICTED: quinone oxidoreductase PIG3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000041_010 |
0.3009798048 |
- |
- |
pfkB-type carbohydrate kinase family protein [Populus trichocarpa] |
| 11 |
Hb_002874_040 |
0.3011547401 |
- |
- |
- |
| 12 |
Hb_002304_070 |
0.3034262753 |
- |
- |
- |
| 13 |
Hb_044187_010 |
0.304992056 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 14 |
Hb_000703_270 |
0.3064015186 |
- |
- |
hypothetical protein B456_012G039600 [Gossypium raimondii] |
| 15 |
Hb_002419_030 |
0.3064649984 |
- |
- |
- |
| 16 |
Hb_032705_020 |
0.3077162166 |
- |
- |
PREDICTED: uncharacterized protein LOC104105697 [Nicotiana tomentosiformis] |
| 17 |
Hb_003206_020 |
0.3080146241 |
- |
- |
PREDICTED: putative vacuolar protein sorting-associated protein 13A isoform X2 [Vitis vinifera] |
| 18 |
Hb_000286_200 |
0.3125506626 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_001278_070 |
0.3140139702 |
- |
- |
hypothetical protein glysoja_020139 [Glycine soja] |
| 20 |
Hb_182831_010 |
0.3142267257 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |