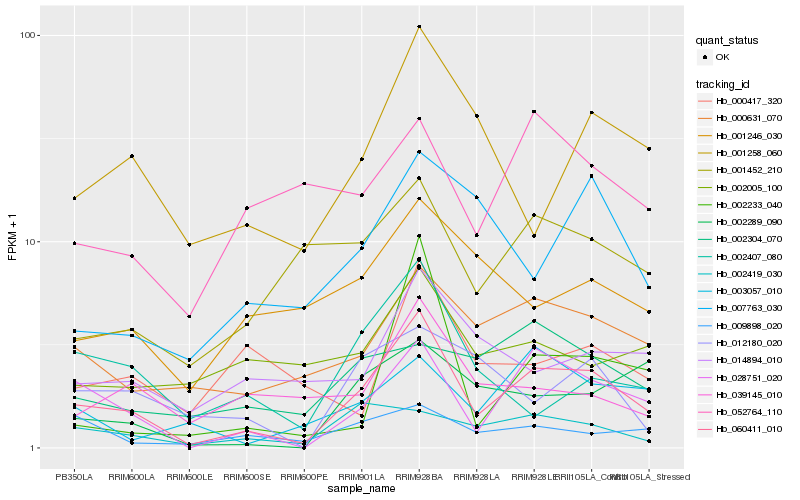
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_060411_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC100246093 [Vitis vinifera] |
| 2 |
Hb_028751_020 |
0.1801276684 |
- |
- |
PREDICTED: SCY1-like protein 2 [Jatropha curcas] |
| 3 |
Hb_000631_070 |
0.2255986097 |
transcription factor |
TF Family: C2H2 |
PREDICTED: histone deacetylase 5 [Jatropha curcas] |
| 4 |
Hb_002419_030 |
0.2535190106 |
- |
- |
- |
| 5 |
Hb_001258_060 |
0.2560581535 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxykinase [ATP] [Jatropha curcas] |
| 6 |
Hb_001452_210 |
0.2590282448 |
- |
- |
molybdopterin biosynthesis CNX1 protein [Arabidopsis thaliana] |
| 7 |
Hb_002407_080 |
0.2606387891 |
- |
- |
hypothetical protein JCGZ_00752 [Jatropha curcas] |
| 8 |
Hb_002289_090 |
0.2614361799 |
- |
- |
cytoplasmic tRNA 2-thiolation protein 2-like [Scleropages formosus] |
| 9 |
Hb_014894_010 |
0.2614641672 |
- |
- |
hypothetical protein CICLE_v10001869mg [Citrus clementina] |
| 10 |
Hb_039145_010 |
0.2628681999 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 11 |
Hb_009898_020 |
0.2638139169 |
- |
- |
hypothetical protein JCGZ_23951 [Jatropha curcas] |
| 12 |
Hb_003057_010 |
0.2665673037 |
- |
- |
hypothetical protein JCGZ_06666 [Jatropha curcas] |
| 13 |
Hb_001246_030 |
0.2689511788 |
- |
- |
hypothetical protein JCGZ_14672 [Jatropha curcas] |
| 14 |
Hb_002233_040 |
0.2721291787 |
- |
- |
PREDICTED: uncharacterized protein LOC105629659 [Jatropha curcas] |
| 15 |
Hb_002304_070 |
0.272207721 |
- |
- |
- |
| 16 |
Hb_007763_030 |
0.2738847337 |
- |
- |
PREDICTED: uncharacterized protein LOC105638154 [Jatropha curcas] |
| 17 |
Hb_012180_020 |
0.275193709 |
- |
- |
hypothetical protein POPTR_0001s15100g [Populus trichocarpa] |
| 18 |
Hb_052764_110 |
0.276205839 |
- |
- |
PREDICTED: quinone oxidoreductase PIG3 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000417_320 |
0.2769962169 |
- |
- |
hypothetical protein CICLE_v10000333mg [Citrus clementina] |
| 20 |
Hb_002005_100 |
0.2774001946 |
- |
- |
- |