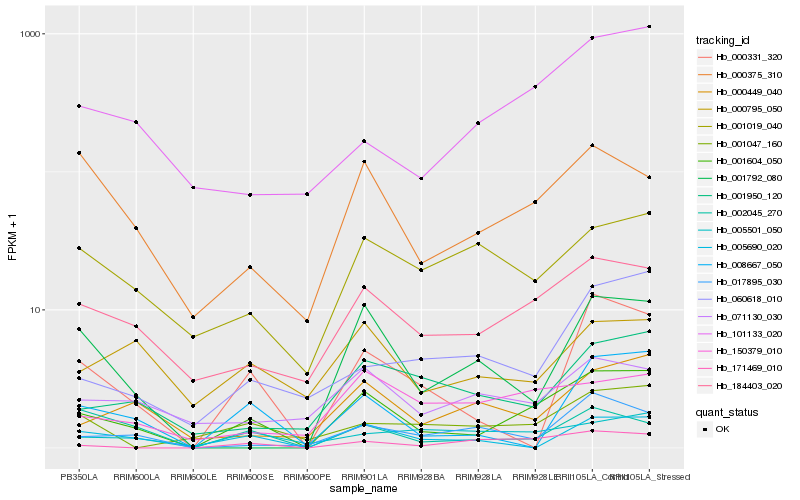
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001604_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639208 [Jatropha curcas] |
| 2 |
Hb_184403_020 |
0.2014325661 |
- |
- |
PREDICTED: pre-rRNA-processing protein ESF2 [Vitis vinifera] |
| 3 |
Hb_000375_310 |
0.2397037882 |
- |
- |
hypothetical protein JCGZ_18643 [Jatropha curcas] |
| 4 |
Hb_171469_010 |
0.2455486103 |
- |
- |
PREDICTED: uncharacterized protein LOC105775124 [Gossypium raimondii] |
| 5 |
Hb_001950_120 |
0.2461142397 |
- |
- |
PREDICTED: probable cadmium/zinc-transporting ATPase HMA1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_101133_020 |
0.2480179605 |
- |
- |
PREDICTED: 15.7 kDa heat shock protein, peroxisomal [Jatropha curcas] |
| 7 |
Hb_000449_040 |
0.248916164 |
- |
- |
protein with unknown function [Ricinus communis] |
| 8 |
Hb_008667_050 |
0.2496229252 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Malus domestica] |
| 9 |
Hb_000331_320 |
0.2513952221 |
- |
- |
- |
| 10 |
Hb_150379_010 |
0.2521366212 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647563 [Jatropha curcas] |
| 11 |
Hb_005501_050 |
0.259426866 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 12 |
Hb_060618_010 |
0.2598674073 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 13 |
Hb_005690_020 |
0.2602946853 |
- |
- |
hypothetical protein POPTR_0006s08200g [Populus trichocarpa] |
| 14 |
Hb_017895_030 |
0.2621537994 |
transcription factor |
TF Family: C2H2 |
- |
| 15 |
Hb_000795_050 |
0.2629353202 |
- |
- |
hypothetical protein CISIN_1g0154952mg, partial [Citrus sinensis] |
| 16 |
Hb_001792_080 |
0.2637949596 |
- |
- |
unknown [Populus trichocarpa] |
| 17 |
Hb_002045_270 |
0.2639769385 |
- |
- |
hypothetical protein JCGZ_21864 [Jatropha curcas] |
| 18 |
Hb_071130_030 |
0.2643140257 |
- |
- |
PREDICTED: dnaJ homolog subfamily C member 17 [Jatropha curcas] |
| 19 |
Hb_001019_040 |
0.2653125186 |
- |
- |
PREDICTED: monothiol glutaredoxin-S7, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001047_160 |
0.2659986555 |
- |
- |
- |