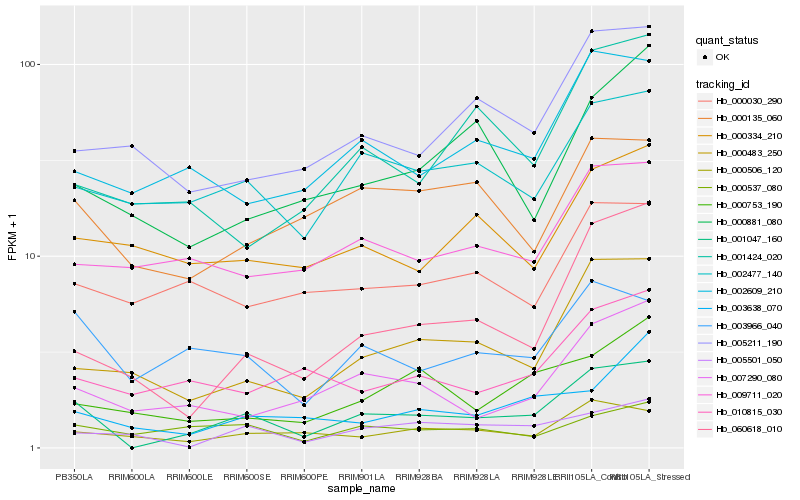
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001047_160 |
0.0 |
- |
- |
- |
| 2 |
Hb_000483_250 |
0.1976691106 |
- |
- |
PREDICTED: uncharacterized protein LOC105803540 [Gossypium raimondii] |
| 3 |
Hb_060618_010 |
0.1977093016 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 4 |
Hb_003966_040 |
0.2055947455 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2 [Jatropha curcas] |
| 5 |
Hb_002477_140 |
0.2061909794 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-3 isoform X1 [Jatropha curcas] |
| 6 |
Hb_007290_080 |
0.2071719136 |
- |
- |
hypothetical protein CISIN_1g047098mg [Citrus sinensis] |
| 7 |
Hb_002609_210 |
0.2090583971 |
- |
- |
PREDICTED: pleckstrin homology domain-containing protein 1 [Jatropha curcas] |
| 8 |
Hb_000506_120 |
0.2171711664 |
- |
- |
hypothetical protein AALP_AA6G154200 [Arabis alpina] |
| 9 |
Hb_005211_190 |
0.2192383141 |
- |
- |
PREDICTED: uncharacterized protein LOC105641994 [Jatropha curcas] |
| 10 |
Hb_000135_060 |
0.2201615951 |
transcription factor |
TF Family: MED6 |
PREDICTED: mediator of RNA polymerase II transcription subunit 6 [Jatropha curcas] |
| 11 |
Hb_010815_030 |
0.2228316714 |
- |
- |
PREDICTED: (-)-germacrene D synthase [Vitis vinifera] |
| 12 |
Hb_001424_020 |
0.2230901727 |
- |
- |
PREDICTED: AP-1 complex subunit sigma-2 isoform X2 [Solanum lycopersicum] |
| 13 |
Hb_009711_020 |
0.2233632484 |
- |
- |
PREDICTED: uncharacterized protein LOC105647794 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000753_190 |
0.224388316 |
- |
- |
PREDICTED: D-aminoacyl-tRNA deacylase [Populus euphratica] |
| 15 |
Hb_003638_070 |
0.2264534639 |
- |
- |
hypothetical protein EUGRSUZ_E00638 [Eucalyptus grandis] |
| 16 |
Hb_000881_080 |
0.2265333976 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_005501_050 |
0.2280376484 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 18 |
Hb_000537_080 |
0.2292367969 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000030_290 |
0.2304183822 |
- |
- |
PREDICTED: uncharacterized protein LOC100247068 [Vitis vinifera] |
| 20 |
Hb_000334_210 |
0.2318460225 |
- |
- |
PREDICTED: uncharacterized protein LOC105628719 isoform X1 [Jatropha curcas] |