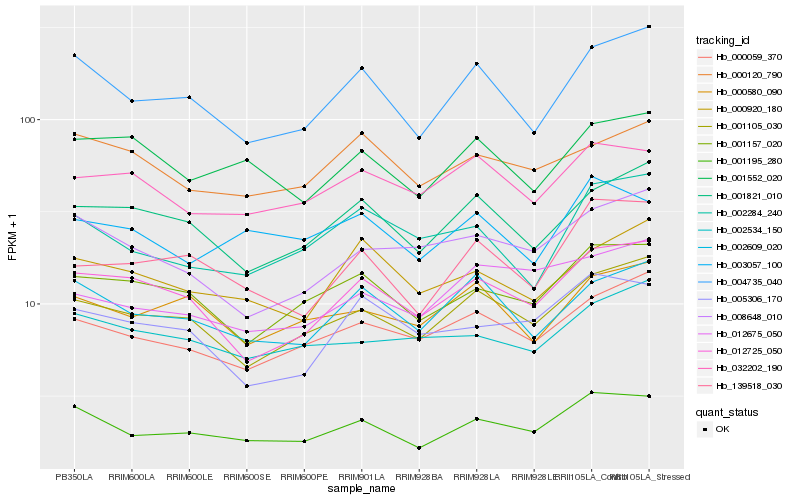
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001195_280 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103928873 isoform X1 [Pyrus x bretschneideri] |
| 2 |
Hb_004735_040 |
0.0723890026 |
- |
- |
PREDICTED: GTP-binding nuclear protein Ran1A-like [Jatropha curcas] |
| 3 |
Hb_001105_030 |
0.0763658875 |
- |
- |
PREDICTED: probable caffeoyl-CoA O-methyltransferase At4g26220 [Jatropha curcas] |
| 4 |
Hb_001157_020 |
0.0769725839 |
- |
- |
PREDICTED: uncharacterized protein LOC105649844 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002609_020 |
0.0847319552 |
- |
- |
PREDICTED: phosphatidylinositol N-acetylglucosaminyltransferase subunit P-like [Jatropha curcas] |
| 6 |
Hb_000059_370 |
0.0855451004 |
- |
- |
PREDICTED: U6 snRNA phosphodiesterase [Jatropha curcas] |
| 7 |
Hb_012725_050 |
0.0884408379 |
transcription factor |
TF Family: SET |
set domain protein, putative [Ricinus communis] |
| 8 |
Hb_003057_100 |
0.0933750024 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: S-adenosylmethionine mitochondrial carrier protein-like [Malus domestica] |
| 9 |
Hb_139518_030 |
0.0946426438 |
- |
- |
protein kinase atn1, putative [Ricinus communis] |
| 10 |
Hb_001821_010 |
0.0950057978 |
- |
- |
PREDICTED: uncharacterized protein C9orf78 [Jatropha curcas] |
| 11 |
Hb_000580_090 |
0.095381883 |
- |
- |
PREDICTED: tRNA 2'-phosphotransferase 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002534_150 |
0.0957055845 |
- |
- |
catalytic, putative [Ricinus communis] |
| 13 |
Hb_001552_020 |
0.095941723 |
- |
- |
PREDICTED: uncharacterized protein LOC105639653 [Jatropha curcas] |
| 14 |
Hb_032202_190 |
0.0975375131 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 21 [Gossypium raimondii] |
| 15 |
Hb_000120_790 |
0.0982765634 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein H isoform X1 [Jatropha curcas] |
| 16 |
Hb_000920_180 |
0.1000089381 |
- |
- |
PREDICTED: F-box protein SKIP5 [Jatropha curcas] |
| 17 |
Hb_008648_010 |
0.100453222 |
- |
- |
PREDICTED: pre-rRNA-processing protein ESF2 [Jatropha curcas] |
| 18 |
Hb_002284_240 |
0.1005722118 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 115 isoform X3 [Jatropha curcas] |
| 19 |
Hb_012675_050 |
0.1012144738 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_005306_170 |
0.1012746555 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 4-like [Jatropha curcas] |