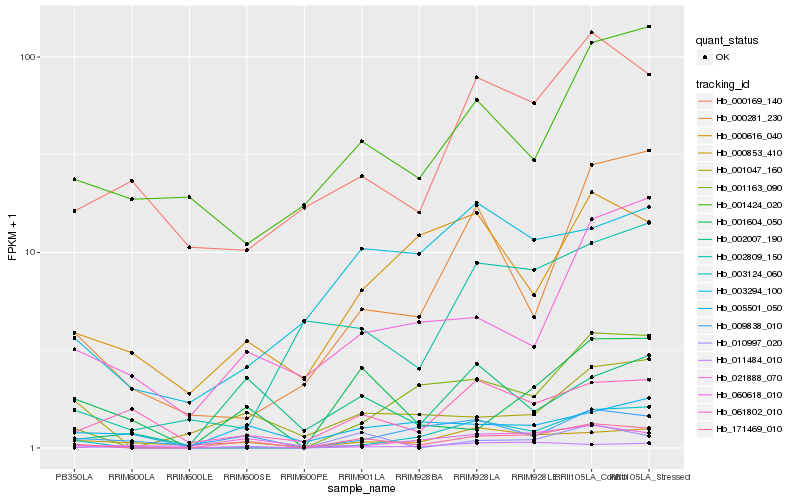
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_171469_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105775124 [Gossypium raimondii] |
| 2 |
Hb_001604_050 |
0.2455486103 |
- |
- |
PREDICTED: uncharacterized protein LOC105639208 [Jatropha curcas] |
| 3 |
Hb_001047_160 |
0.2518301228 |
- |
- |
- |
| 4 |
Hb_021888_070 |
0.2582224393 |
- |
- |
Putative CCR4-associated factor 1 like 11 [Glycine soja] |
| 5 |
Hb_009838_010 |
0.2626579893 |
- |
- |
- |
| 6 |
Hb_000853_410 |
0.2679595003 |
- |
- |
- |
| 7 |
Hb_000169_140 |
0.2685261354 |
- |
- |
PREDICTED: uncharacterized protein LOC105643397 [Jatropha curcas] |
| 8 |
Hb_002007_190 |
0.2699036311 |
- |
- |
hypothetical protein RCOM_1432220 [Ricinus communis] |
| 9 |
Hb_001163_090 |
0.2719419944 |
- |
- |
hypothetical protein POPTR_0009s14920g [Populus trichocarpa] |
| 10 |
Hb_003124_060 |
0.2723412283 |
- |
- |
Beta-ureidopropionase, putative [Ricinus communis] |
| 11 |
Hb_010997_020 |
0.273925449 |
- |
- |
- |
| 12 |
Hb_005501_050 |
0.2774731625 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 13 |
Hb_060618_010 |
0.278320333 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 14 |
Hb_003294_100 |
0.2832132981 |
- |
- |
hypothetical protein VITISV_009629 [Vitis vinifera] |
| 15 |
Hb_000616_040 |
0.2835222421 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 16 |
Hb_061802_010 |
0.2837816869 |
- |
- |
hypothetical protein VITISV_011986 [Vitis vinifera] |
| 17 |
Hb_002809_150 |
0.2846778928 |
transcription factor |
TF Family: M-type |
hypothetical protein CISIN_1g0283632mg, partial [Citrus sinensis] |
| 18 |
Hb_011484_010 |
0.2850972679 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 19 |
Hb_000281_230 |
0.2855267794 |
- |
- |
PREDICTED: probable lysine--tRNA ligase, cytoplasmic isoform X2 [Jatropha curcas] |
| 20 |
Hb_001424_020 |
0.2884065214 |
- |
- |
PREDICTED: AP-1 complex subunit sigma-2 isoform X2 [Solanum lycopersicum] |