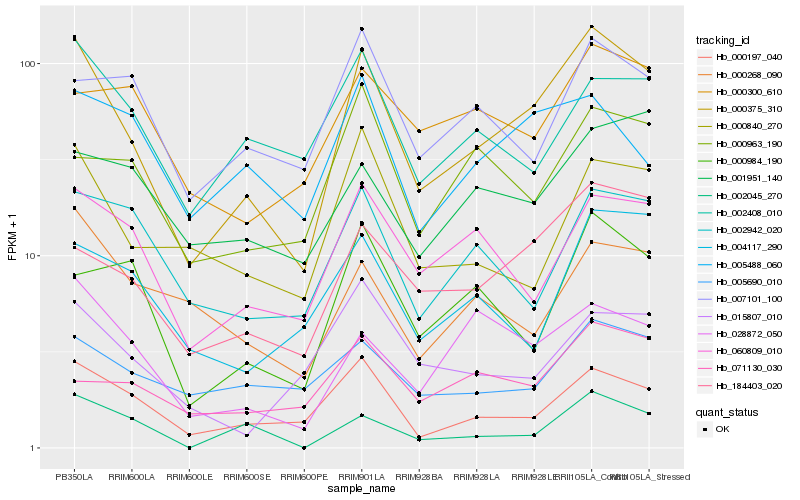
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000375_310 |
0.0 |
- |
- |
hypothetical protein JCGZ_18643 [Jatropha curcas] |
| 2 |
Hb_000197_040 |
0.1486194811 |
- |
- |
AMP-activated protein kinase, gamma regulatory subunit, putative [Ricinus communis] |
| 3 |
Hb_184403_020 |
0.1520906134 |
- |
- |
PREDICTED: pre-rRNA-processing protein ESF2 [Vitis vinifera] |
| 4 |
Hb_060809_010 |
0.173513334 |
- |
- |
PREDICTED: uncharacterized protein LOC105163892 [Sesamum indicum] |
| 5 |
Hb_028872_050 |
0.1784105142 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial [Jatropha curcas] |
| 6 |
Hb_000840_270 |
0.179022952 |
- |
- |
PREDICTED: small RNA degrading nuclease 1 [Jatropha curcas] |
| 7 |
Hb_005690_010 |
0.1836787426 |
- |
- |
PREDICTED: origin recognition complex subunit 2 [Populus euphratica] |
| 8 |
Hb_002408_010 |
0.1868607461 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 9 |
Hb_002942_020 |
0.1883746255 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 10 |
Hb_004117_290 |
0.1887142149 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000984_190 |
0.1890280533 |
- |
- |
- |
| 12 |
Hb_000268_090 |
0.193111776 |
- |
- |
PREDICTED: uncharacterized protein LOC105628378 isoform X2 [Jatropha curcas] |
| 13 |
Hb_015807_010 |
0.1940279566 |
- |
- |
hypothetical protein POPTR_0001s44430g [Populus trichocarpa] |
| 14 |
Hb_000963_190 |
0.1958388356 |
- |
- |
Glutamine amidotransferase subunit pdxT, putative [Ricinus communis] |
| 15 |
Hb_071130_030 |
0.1964066194 |
- |
- |
PREDICTED: dnaJ homolog subfamily C member 17 [Jatropha curcas] |
| 16 |
Hb_002045_270 |
0.1964465128 |
- |
- |
hypothetical protein JCGZ_21864 [Jatropha curcas] |
| 17 |
Hb_007101_100 |
0.1973576156 |
- |
- |
PREDICTED: protein LSM12 homolog A-like [Jatropha curcas] |
| 18 |
Hb_000300_610 |
0.1974654451 |
- |
- |
cytosolic copper/zinc superoxide dismutase [Hevea brasiliensis] |
| 19 |
Hb_001951_140 |
0.1981167043 |
- |
- |
PREDICTED: uncharacterized protein LOC105649757 isoform X1 [Jatropha curcas] |
| 20 |
Hb_005488_060 |
0.1982639726 |
- |
- |
cytochrome P450, putative [Ricinus communis] |