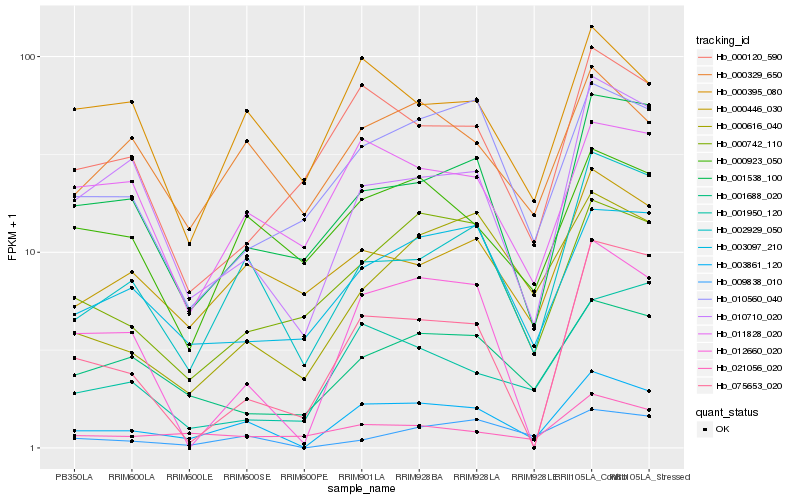
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003861_120 |
0.0 |
transcription factor |
TF Family: RWP-RK |
transcription factor, putative [Ricinus communis] |
| 2 |
Hb_002929_050 |
0.1403707337 |
- |
- |
PREDICTED: polyadenylate-binding protein 6 isoform X1 [Jatropha curcas] |
| 3 |
Hb_010710_020 |
0.1754401825 |
- |
- |
PREDICTED: uncharacterized protein LOC103322096 [Prunus mume] |
| 4 |
Hb_009838_010 |
0.1771844664 |
- |
- |
- |
| 5 |
Hb_001538_100 |
0.1776232773 |
- |
- |
PREDICTED: histone deacetylase 6 [Jatropha curcas] |
| 6 |
Hb_003097_210 |
0.1797793217 |
- |
- |
PREDICTED: uncharacterized protein LOC105631194 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000446_030 |
0.1853086176 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 8 |
Hb_012660_020 |
0.186737111 |
- |
- |
- |
| 9 |
Hb_000742_110 |
0.1871589115 |
- |
- |
hypothetical protein JCGZ_09215 [Jatropha curcas] |
| 10 |
Hb_001688_020 |
0.1878938777 |
- |
- |
PREDICTED: uncharacterized protein LOC103438026 [Malus domestica] |
| 11 |
Hb_000329_650 |
0.1887100576 |
- |
- |
hypothetical protein POPTR_0014s056001g, partial [Populus trichocarpa] |
| 12 |
Hb_010560_040 |
0.1897867656 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 11 [Jatropha curcas] |
| 13 |
Hb_000616_040 |
0.1898654917 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 14 |
Hb_000923_050 |
0.1922757277 |
- |
- |
PREDICTED: transcription factor bHLH113 isoform X1 [Jatropha curcas] |
| 15 |
Hb_011828_020 |
0.1933128025 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-10-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_075653_020 |
0.1937898021 |
- |
- |
- |
| 17 |
Hb_021056_020 |
0.1939978638 |
- |
- |
plasma membrane H+-ATPase [Hevea brasiliensis] |
| 18 |
Hb_000395_080 |
0.1967852889 |
- |
- |
PREDICTED: uncharacterized protein LOC105645573 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001950_120 |
0.197449693 |
- |
- |
PREDICTED: probable cadmium/zinc-transporting ATPase HMA1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000120_590 |
0.1974877813 |
- |
- |
PREDICTED: uncharacterized protein LOC105116418 [Populus euphratica] |