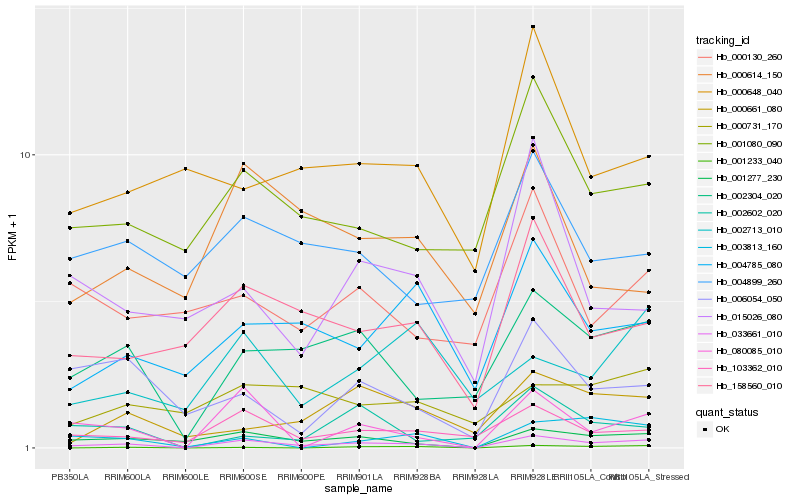
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033661_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105852393, partial [Cicer arietinum] |
| 2 |
Hb_001277_230 |
0.2079768775 |
- |
- |
- |
| 3 |
Hb_002304_020 |
0.2146131219 |
- |
- |
- |
| 4 |
Hb_080085_010 |
0.2285872134 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 5 |
Hb_103362_010 |
0.2308369208 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 6 |
Hb_158560_010 |
0.247938059 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable acyl-activating enzyme 1, peroxisomal [Jatropha curcas] |
| 7 |
Hb_004785_080 |
0.2596764503 |
- |
- |
hypothetical protein POPTR_0005s21250g [Populus trichocarpa] |
| 8 |
Hb_001080_090 |
0.2627305272 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g30600 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000661_080 |
0.2709385581 |
- |
- |
monoxygenase, putative [Ricinus communis] |
| 10 |
Hb_006054_050 |
0.2724378618 |
- |
- |
PREDICTED: uncharacterized protein LOC105649250, partial [Jatropha curcas] |
| 11 |
Hb_003813_160 |
0.2754734314 |
- |
- |
- |
| 12 |
Hb_001233_040 |
0.2797432374 |
- |
- |
PREDICTED: uncharacterized protein LOC105775263 [Gossypium raimondii] |
| 13 |
Hb_015026_080 |
0.2813381203 |
- |
- |
- |
| 14 |
Hb_002713_010 |
0.2822125547 |
- |
- |
multidrug resistance protein 1, 2, putative [Ricinus communis] |
| 15 |
Hb_000731_170 |
0.28258743 |
- |
- |
PREDICTED: phospholipase A I [Jatropha curcas] |
| 16 |
Hb_002602_020 |
0.2857885009 |
- |
- |
- |
| 17 |
Hb_000648_040 |
0.2863636543 |
- |
- |
unknown [Populus trichocarpa] |
| 18 |
Hb_000614_150 |
0.2884672511 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory ankyrin repeat subunit B-like [Jatropha curcas] |
| 19 |
Hb_004899_260 |
0.2893368825 |
- |
- |
PREDICTED: uncharacterized protein LOC105638006 [Jatropha curcas] |
| 20 |
Hb_000130_260 |
0.2897245912 |
rubber biosynthesis |
Gene Name: Dimethyallyltransferase |
PREDICTED: tRNA dimethylallyltransferase 9 isoform X1 [Jatropha curcas] |