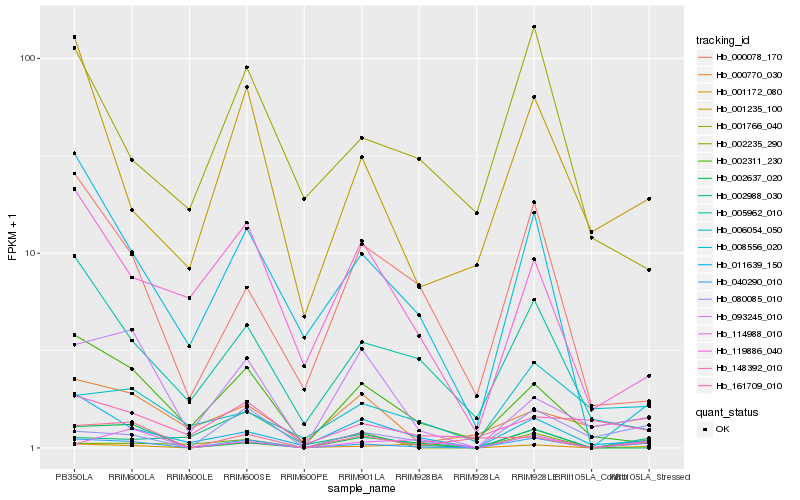
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_040290_010 |
0.0 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 2 |
Hb_080085_010 |
0.2464519017 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 3 |
Hb_001172_080 |
0.2889979084 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 4 |
Hb_011639_150 |
0.2964238919 |
- |
- |
- |
| 5 |
Hb_161709_010 |
0.3009670784 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 6 |
Hb_001235_100 |
0.301736456 |
- |
- |
heat-shock protein, putative [Ricinus communis] |
| 7 |
Hb_093245_010 |
0.3080125098 |
- |
- |
- |
| 8 |
Hb_002311_230 |
0.308190861 |
- |
- |
- |
| 9 |
Hb_008556_020 |
0.3096594107 |
- |
- |
- |
| 10 |
Hb_114988_010 |
0.3106761144 |
- |
- |
- |
| 11 |
Hb_000078_170 |
0.318631388 |
- |
- |
- |
| 12 |
Hb_005962_010 |
0.3233218688 |
- |
- |
- |
| 13 |
Hb_002637_020 |
0.3267909853 |
- |
- |
- |
| 14 |
Hb_000770_030 |
0.3281709155 |
- |
- |
- |
| 15 |
Hb_006054_050 |
0.329643662 |
- |
- |
PREDICTED: uncharacterized protein LOC105649250, partial [Jatropha curcas] |
| 16 |
Hb_119886_040 |
0.3304440914 |
- |
- |
- |
| 17 |
Hb_002235_290 |
0.3339265648 |
- |
- |
PREDICTED: casparian strip membrane protein 5 [Jatropha curcas] |
| 18 |
Hb_002988_030 |
0.3367321965 |
- |
- |
hypothetical protein EUGRSUZ_K02073 [Eucalyptus grandis] |
| 19 |
Hb_001766_040 |
0.3367384231 |
- |
- |
chaperone clpb, putative [Ricinus communis] |
| 20 |
Hb_148392_010 |
0.339345861 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |