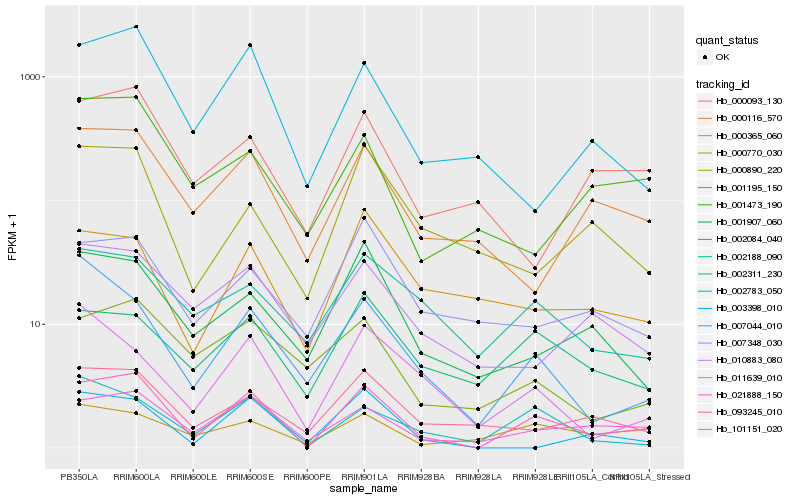
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_093245_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_021888_150 |
0.1846768364 |
- |
- |
- |
| 3 |
Hb_002311_230 |
0.2137118591 |
- |
- |
- |
| 4 |
Hb_101151_020 |
0.2260256435 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_000770_030 |
0.2260721441 |
- |
- |
- |
| 6 |
Hb_002783_050 |
0.2288298296 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 7 |
Hb_000116_570 |
0.2361328507 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 5-like [Jatropha curcas] |
| 8 |
Hb_007044_010 |
0.2364390717 |
- |
- |
- |
| 9 |
Hb_002084_040 |
0.2377165821 |
- |
- |
PREDICTED: DNA-directed RNA polymerase 3, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001907_060 |
0.239882758 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 5 [Jatropha curcas] |
| 11 |
Hb_003398_010 |
0.2399688839 |
- |
- |
PREDICTED: uncharacterized protein At4g02000-like [Eucalyptus grandis] |
| 12 |
Hb_000890_220 |
0.2432072958 |
transcription factor |
TF Family: SNF2 |
PREDICTED: putative chromatin-remodeling complex ATPase chain isoform X1 [Jatropha curcas] |
| 13 |
Hb_011639_010 |
0.2438257859 |
- |
- |
- |
| 14 |
Hb_001195_150 |
0.2452751424 |
transcription factor |
TF Family: MYB-related |
myb family transcription factor family protein [Populus trichocarpa] |
| 15 |
Hb_000093_130 |
0.2454762369 |
- |
- |
glycine-rich RNA-binding protein, putative [Ricinus communis] |
| 16 |
Hb_010883_080 |
0.2462292153 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHR1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000365_060 |
0.2467659032 |
- |
- |
5'->3' exoribonuclease, putative [Ricinus communis] |
| 18 |
Hb_001473_190 |
0.2468147656 |
- |
- |
50S ribosomal protein L20 [Hevea brasiliensis] |
| 19 |
Hb_007348_030 |
0.2473509298 |
- |
- |
PREDICTED: uncharacterized protein LOC105645596 isoform X2 [Jatropha curcas] |
| 20 |
Hb_002188_090 |
0.2482908281 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40A isoform X1 [Jatropha curcas] |