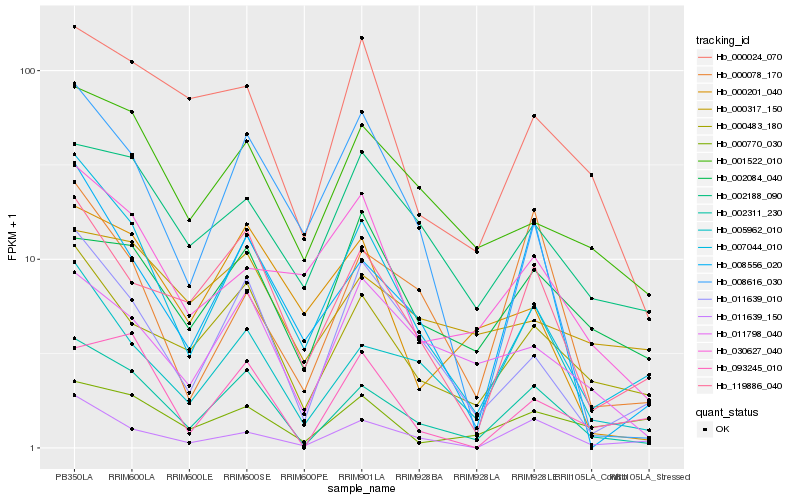
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002311_230 |
0.0 |
- |
- |
- |
| 2 |
Hb_005962_010 |
0.149546504 |
- |
- |
- |
| 3 |
Hb_000483_180 |
0.157935223 |
- |
- |
- |
| 4 |
Hb_119886_040 |
0.1659074906 |
- |
- |
- |
| 5 |
Hb_007044_010 |
0.170145015 |
- |
- |
- |
| 6 |
Hb_008556_020 |
0.1780418776 |
- |
- |
- |
| 7 |
Hb_011639_010 |
0.1786207432 |
- |
- |
- |
| 8 |
Hb_011639_150 |
0.1953417585 |
- |
- |
- |
| 9 |
Hb_001522_010 |
0.202397614 |
- |
- |
PREDICTED: nucleolar protein 14 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000078_170 |
0.203161629 |
- |
- |
- |
| 11 |
Hb_000024_070 |
0.2042577959 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 6-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_011798_040 |
0.2054515498 |
- |
- |
hypothetical protein ZEAMMB73_778895 [Zea mays] |
| 13 |
Hb_093245_010 |
0.2137118591 |
- |
- |
- |
| 14 |
Hb_002188_090 |
0.2149019542 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40A isoform X1 [Jatropha curcas] |
| 15 |
Hb_002084_040 |
0.2239897251 |
- |
- |
PREDICTED: DNA-directed RNA polymerase 3, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000201_040 |
0.2272234124 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Populus euphratica] |
| 17 |
Hb_008616_030 |
0.2279274285 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-10 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000317_150 |
0.2357609043 |
- |
- |
PREDICTED: uncharacterized protein LOC104901272 [Beta vulgaris subsp. vulgaris] |
| 19 |
Hb_030627_040 |
0.2357899071 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g59700 [Jatropha curcas] |
| 20 |
Hb_000770_030 |
0.2364496863 |
- |
- |
- |