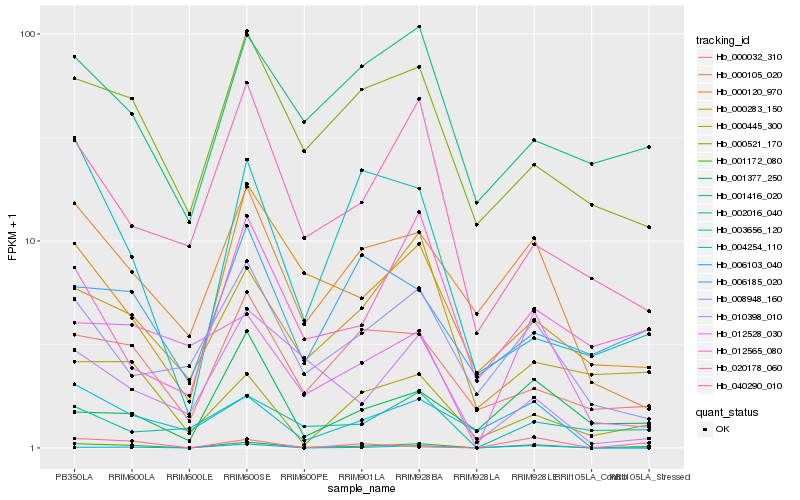
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001172_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 2 |
Hb_000445_300 |
0.2624216828 |
- |
- |
PREDICTED: wall-associated receptor kinase 5-like [Eucalyptus grandis] |
| 3 |
Hb_000032_310 |
0.2704492874 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 4 |
Hb_000105_020 |
0.2708674029 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOR-like [Malus domestica] |
| 5 |
Hb_012565_080 |
0.273917793 |
- |
- |
serine/threonine kinase, putative [Ricinus communis] |
| 6 |
Hb_000521_170 |
0.2774245019 |
- |
- |
PREDICTED: callose synthase 1 [Jatropha curcas] |
| 7 |
Hb_000120_970 |
0.2790786864 |
- |
- |
PREDICTED: probable acyl-activating enzyme 2 isoform X2 [Jatropha curcas] |
| 8 |
Hb_001377_250 |
0.2803870089 |
- |
- |
dynamin, putative [Ricinus communis] |
| 9 |
Hb_001416_020 |
0.28158356 |
- |
- |
H(\+)-transporting atpase plant/fungi plasma membrane type, putative [Ricinus communis] |
| 10 |
Hb_008948_160 |
0.2825830435 |
- |
- |
PREDICTED: uncharacterized protein LOC104214688 isoform X2 [Nicotiana sylvestris] |
| 11 |
Hb_000283_150 |
0.282969855 |
- |
- |
Ankyrin repeat family protein [Theobroma cacao] |
| 12 |
Hb_002016_040 |
0.2845317874 |
rubber biosynthesis |
Gene Name: SRPP8 |
PREDICTED: stress-related protein [Jatropha curcas] |
| 13 |
Hb_020178_060 |
0.2888247649 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 3 [Jatropha curcas] |
| 14 |
Hb_040290_010 |
0.2889979084 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 15 |
Hb_010398_010 |
0.2922197526 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: mediator of RNA polymerase II transcription subunit 15a-like [Malus domestica] |
| 16 |
Hb_004254_110 |
0.2923781044 |
- |
- |
- |
| 17 |
Hb_006185_020 |
0.2924763084 |
- |
- |
PREDICTED: midasin [Jatropha curcas] |
| 18 |
Hb_012528_030 |
0.2943525148 |
- |
- |
- |
| 19 |
Hb_003656_120 |
0.2985371779 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-3-like [Jatropha curcas] |
| 20 |
Hb_006103_040 |
0.2994945391 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |