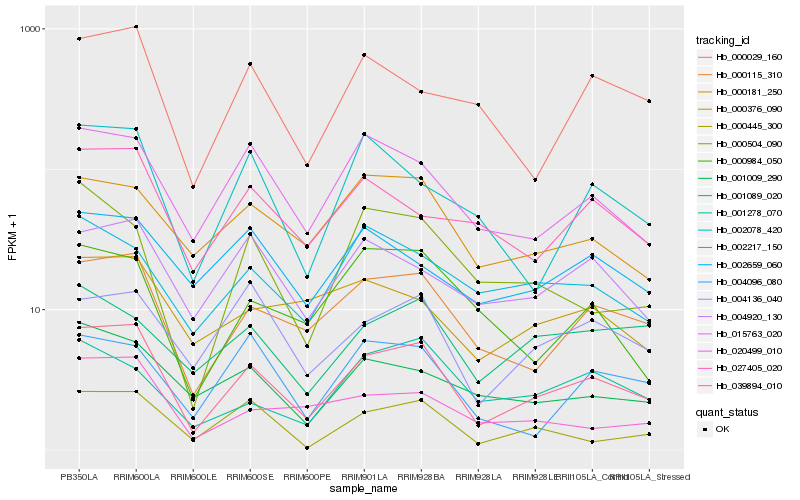
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_039894_010 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_000115_310 |
0.1160118748 |
- |
- |
PREDICTED: zinc finger protein 6 [Jatropha curcas] |
| 3 |
Hb_015763_020 |
0.1527643006 |
- |
- |
PREDICTED: uncharacterized protein LOC105637166 [Jatropha curcas] |
| 4 |
Hb_000445_300 |
0.1606210141 |
- |
- |
PREDICTED: wall-associated receptor kinase 5-like [Eucalyptus grandis] |
| 5 |
Hb_001009_290 |
0.166574209 |
- |
- |
PREDICTED: elongator complex protein 3-like [Populus euphratica] |
| 6 |
Hb_004136_040 |
0.1670275242 |
- |
- |
putative protein kinase [Arabidopsis thaliana] |
| 7 |
Hb_004920_130 |
0.1692765715 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_000181_250 |
0.1704094845 |
- |
- |
PREDICTED: probable bifunctional methylthioribulose-1-phosphate dehydratase/enolase-phosphatase E1 [Jatropha curcas] |
| 9 |
Hb_002078_420 |
0.1709293841 |
- |
- |
PREDICTED: F-box only protein 6 [Jatropha curcas] |
| 10 |
Hb_027405_020 |
0.1709583096 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase XBAT31 [Jatropha curcas] |
| 11 |
Hb_020499_010 |
0.1709975986 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 12 |
Hb_000029_160 |
0.1731494264 |
- |
- |
Nitrilase, putative [Ricinus communis] |
| 13 |
Hb_004096_080 |
0.1775775836 |
- |
- |
PREDICTED: IST1 homolog [Jatropha curcas] |
| 14 |
Hb_000504_090 |
0.1807698337 |
- |
- |
PREDICTED: titin isoform X10 [Jatropha curcas] |
| 15 |
Hb_002217_150 |
0.1814591416 |
- |
- |
PREDICTED: nuclear export mediator factor Nemf [Jatropha curcas] |
| 16 |
Hb_000984_050 |
0.1818492968 |
- |
- |
sucrose transporter 5 [Hevea brasiliensis] |
| 17 |
Hb_001089_020 |
0.1825227208 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 2 [Jatropha curcas] |
| 18 |
Hb_002659_060 |
0.1829042645 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3B [Jatropha curcas] |
| 19 |
Hb_000376_090 |
0.183325339 |
- |
- |
Myosin heavy chain, fast skeletal muscle, embryonic, putative [Ricinus communis] |
| 20 |
Hb_001278_070 |
0.1843661418 |
- |
- |
hypothetical protein glysoja_020139 [Glycine soja] |