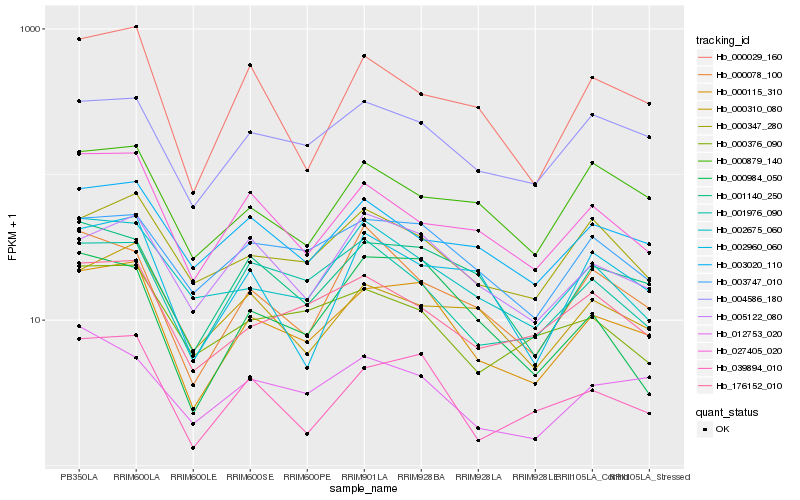
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000115_310 |
0.0 |
- |
- |
PREDICTED: zinc finger protein 6 [Jatropha curcas] |
| 2 |
Hb_039894_010 |
0.1160118748 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_001140_250 |
0.1162945083 |
- |
- |
KDEL motif-containing protein 1 precursor, putative [Ricinus communis] |
| 4 |
Hb_004586_180 |
0.1254026355 |
- |
- |
hypothetical protein JCGZ_15866 [Jatropha curcas] |
| 5 |
Hb_003747_010 |
0.1309123587 |
- |
- |
hypothetical protein JCGZ_24080 [Jatropha curcas] |
| 6 |
Hb_005122_080 |
0.131466227 |
- |
- |
prokaryotic DNA topoisomerase, putative [Ricinus communis] |
| 7 |
Hb_000029_160 |
0.135748768 |
- |
- |
Nitrilase, putative [Ricinus communis] |
| 8 |
Hb_176152_010 |
0.1371082218 |
- |
- |
Myosin heavy chain, fast skeletal muscle, embryonic, putative [Ricinus communis] |
| 9 |
Hb_027405_020 |
0.1376819786 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase XBAT31 [Jatropha curcas] |
| 10 |
Hb_001976_090 |
0.140601902 |
- |
- |
PREDICTED: uncharacterized protein LOC105641696 [Jatropha curcas] |
| 11 |
Hb_000347_280 |
0.140718589 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 7 member B4-like [Populus euphratica] |
| 12 |
Hb_012753_020 |
0.1420675118 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000984_050 |
0.1435633253 |
- |
- |
sucrose transporter 5 [Hevea brasiliensis] |
| 14 |
Hb_000310_080 |
0.1440184014 |
- |
- |
hypothetical protein EUTSA_v10023249mg [Eutrema salsugineum] |
| 15 |
Hb_002675_060 |
0.1470910492 |
- |
- |
PREDICTED: uncharacterized protein LOC105634954 isoform X2 [Jatropha curcas] |
| 16 |
Hb_003020_110 |
0.1479779652 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 1 isoform X2 [Populus euphratica] |
| 17 |
Hb_000078_100 |
0.1485983716 |
- |
- |
PREDICTED: topless-related protein 4-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_002960_060 |
0.1489966463 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 28 [Jatropha curcas] |
| 19 |
Hb_000879_140 |
0.1499385028 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0283697-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_000376_090 |
0.150457815 |
- |
- |
Myosin heavy chain, fast skeletal muscle, embryonic, putative [Ricinus communis] |