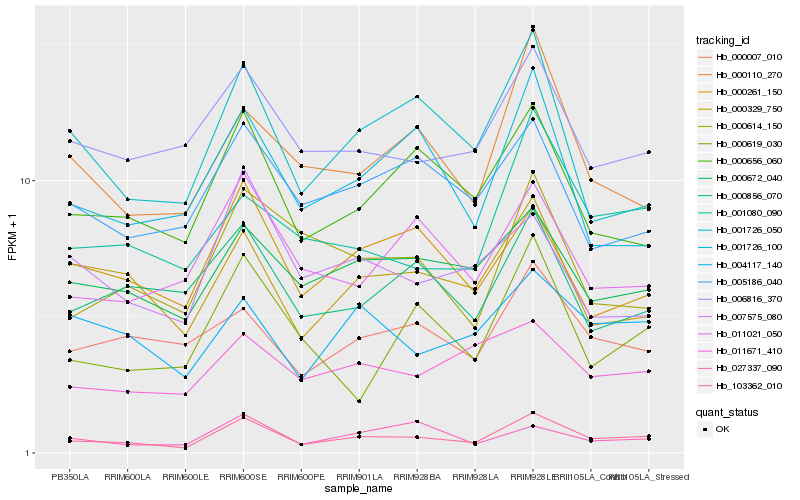
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_103362_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 2 |
Hb_000656_060 |
0.1313253444 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_001726_050 |
0.1399962196 |
transcription factor |
TF Family: C3H |
Nucleic acid binding,zinc ion binding,DNA binding, putative isoform 1 [Theobroma cacao] |
| 4 |
Hb_000110_270 |
0.1403157412 |
- |
- |
prp4, putative [Ricinus communis] |
| 5 |
Hb_000329_750 |
0.1423450341 |
- |
- |
PREDICTED: uncharacterized protein LOC105643205 [Jatropha curcas] |
| 6 |
Hb_011021_050 |
0.1432559227 |
- |
- |
PREDICTED: uncharacterized protein LOC105644387 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000672_040 |
0.1447971835 |
- |
- |
PREDICTED: ENHANCER OF AG-4 protein 2 [Jatropha curcas] |
| 8 |
Hb_000261_150 |
0.1451071369 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001080_090 |
0.1455011435 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g30600 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001726_100 |
0.1462499795 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 19 isoform X2 [Jatropha curcas] |
| 11 |
Hb_006816_370 |
0.1475603759 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105647456 [Jatropha curcas] |
| 12 |
Hb_000007_010 |
0.1511541662 |
- |
- |
PREDICTED: protein PHYLLO, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000619_030 |
0.1515177153 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_004117_140 |
0.1520252915 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g16710, mitochondrial [Populus euphratica] |
| 15 |
Hb_000856_070 |
0.1523809304 |
- |
- |
PREDICTED: formin-like protein 5 [Jatropha curcas] |
| 16 |
Hb_000614_150 |
0.1527630537 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory ankyrin repeat subunit B-like [Jatropha curcas] |
| 17 |
Hb_005186_040 |
0.1536170844 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 13 isoform X2 [Jatropha curcas] |
| 18 |
Hb_027337_090 |
0.1548191545 |
- |
- |
PREDICTED: sodium/hydrogen exchanger 7-like [Malus domestica] |
| 19 |
Hb_011671_410 |
0.1558456546 |
transcription factor |
TF Family: SET |
histone-lysine n-methyltransferase, suvh, putative [Ricinus communis] |
| 20 |
Hb_007575_080 |
0.1599613595 |
- |
- |
PREDICTED: lysine-specific histone demethylase 1 homolog 3 [Jatropha curcas] |