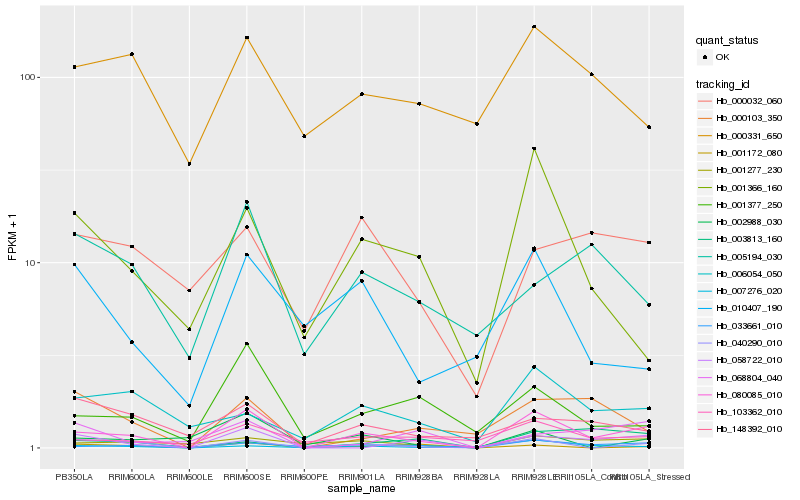
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_080085_010 |
0.0 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 2 |
Hb_033661_010 |
0.2285872134 |
- |
- |
PREDICTED: uncharacterized protein LOC105852393, partial [Cicer arietinum] |
| 3 |
Hb_040290_010 |
0.2464519017 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 4 |
Hb_103362_010 |
0.2795614543 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 5 |
Hb_058722_010 |
0.282598773 |
- |
- |
- |
| 6 |
Hb_001377_250 |
0.2833556091 |
- |
- |
dynamin, putative [Ricinus communis] |
| 7 |
Hb_001277_230 |
0.2886388223 |
- |
- |
- |
| 8 |
Hb_007276_020 |
0.3035194045 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 9 |
Hb_006054_050 |
0.3037310673 |
- |
- |
PREDICTED: uncharacterized protein LOC105649250, partial [Jatropha curcas] |
| 10 |
Hb_001366_160 |
0.306507071 |
- |
- |
- |
| 11 |
Hb_000103_350 |
0.3206734607 |
- |
- |
- |
| 12 |
Hb_002988_030 |
0.3234834046 |
- |
- |
hypothetical protein EUGRSUZ_K02073 [Eucalyptus grandis] |
| 13 |
Hb_148392_010 |
0.324796179 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 14 |
Hb_010407_190 |
0.3263195507 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 15 |
Hb_000331_650 |
0.3276005405 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase-like [Jatropha curcas] |
| 16 |
Hb_000032_060 |
0.3294972193 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_068804_040 |
0.331455594 |
- |
- |
- |
| 18 |
Hb_005194_030 |
0.3323884471 |
- |
- |
PREDICTED: putative formamidase C869.04 isoform X1 [Jatropha curcas] |
| 19 |
Hb_003813_160 |
0.3340204363 |
- |
- |
- |
| 20 |
Hb_001172_080 |
0.3345112239 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |