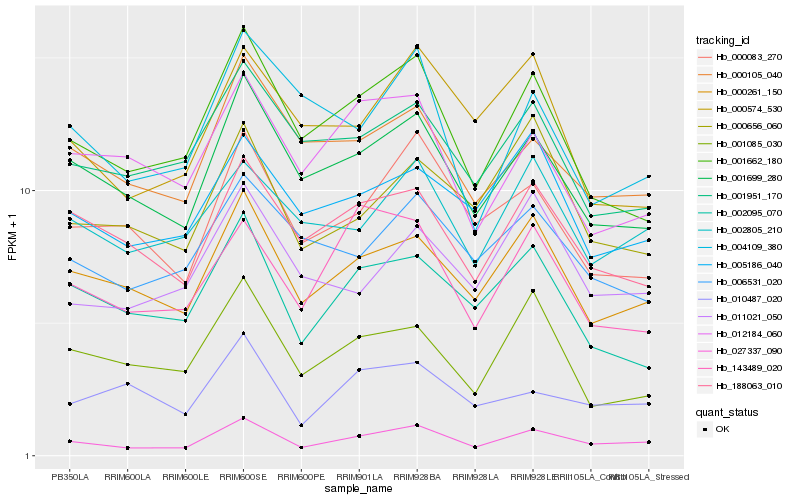
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027337_090 |
0.0 |
- |
- |
PREDICTED: sodium/hydrogen exchanger 7-like [Malus domestica] |
| 2 |
Hb_000261_150 |
0.1038875111 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001699_280 |
0.107683882 |
- |
- |
PREDICTED: transformation/transcription domain-associated protein [Jatropha curcas] |
| 4 |
Hb_001662_180 |
0.111514461 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit 1-like isoform X5 [Citrus sinensis] |
| 5 |
Hb_000083_270 |
0.1233506404 |
- |
- |
PREDICTED: putative uncharacterized protein At4g01020, chloroplastic [Jatropha curcas] |
| 6 |
Hb_188063_010 |
0.1241681774 |
- |
- |
hypothetical protein RCOM_1469330 [Ricinus communis] |
| 7 |
Hb_002095_070 |
0.1298967872 |
- |
- |
PREDICTED: probable pre-mRNA-splicing factor ATP-dependent RNA helicase [Jatropha curcas] |
| 8 |
Hb_000105_040 |
0.1306179912 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOR isoform X2 [Jatropha curcas] |
| 9 |
Hb_000656_060 |
0.1307746987 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 10 |
Hb_143489_020 |
0.1317070081 |
- |
- |
- |
| 11 |
Hb_002805_210 |
0.1318079466 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |
| 12 |
Hb_011021_050 |
0.1322292494 |
- |
- |
PREDICTED: uncharacterized protein LOC105644387 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001085_030 |
0.1334601074 |
- |
- |
Tetratricopeptide repeat-like superfamily protein, putative isoform 1 [Theobroma cacao] |
| 14 |
Hb_005186_040 |
0.1346491368 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 13 isoform X2 [Jatropha curcas] |
| 15 |
Hb_010487_020 |
0.1378803524 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 16 |
Hb_000574_530 |
0.1394671862 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8 [Jatropha curcas] |
| 17 |
Hb_004109_380 |
0.1395950788 |
- |
- |
PREDICTED: callose synthase 9 isoform X4 [Pyrus x bretschneideri] |
| 18 |
Hb_001951_170 |
0.1401019197 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 19 |
Hb_006531_020 |
0.1403606753 |
- |
- |
PREDICTED: autophagy-related protein 13 [Jatropha curcas] |
| 20 |
Hb_012184_060 |
0.1407158361 |
- |
- |
PREDICTED: nuclear pore complex protein NUP1 isoform X1 [Jatropha curcas] |