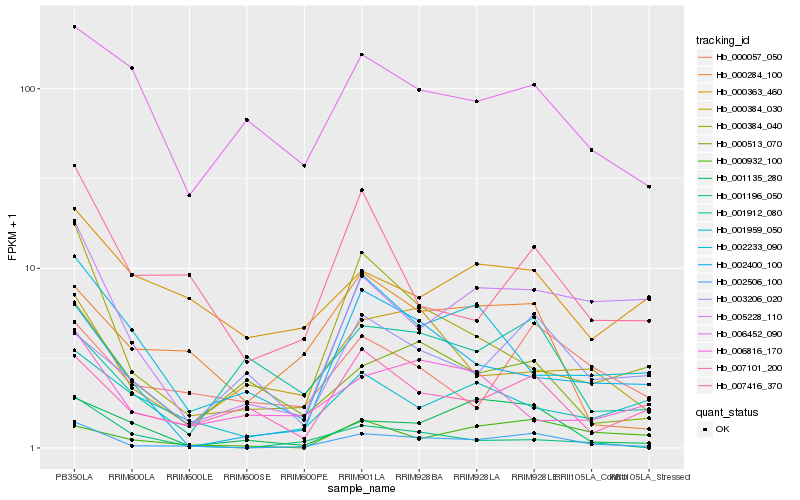
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002506_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641759 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000384_040 |
0.2359066521 |
- |
- |
PREDICTED: uncharacterized protein LOC105640093 [Jatropha curcas] |
| 3 |
Hb_005228_110 |
0.2407626213 |
- |
- |
Cleavage and polyadenylation specificity factor subunit 3 [Fukomys damarensis] |
| 4 |
Hb_007416_370 |
0.2543877769 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_007101_200 |
0.2579933077 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 22 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002400_100 |
0.2582634763 |
- |
- |
PREDICTED: putative UDP-sugar transporter DDB_G0278631 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001196_050 |
0.2589823658 |
- |
- |
- |
| 8 |
Hb_000513_070 |
0.2594711316 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 42 [Jatropha curcas] |
| 9 |
Hb_001135_280 |
0.263082715 |
- |
- |
- |
| 10 |
Hb_000384_030 |
0.2630947214 |
- |
- |
PREDICTED: uncharacterized protein LOC101310565 isoform X1 [Fragaria vesca subsp. vesca] |
| 11 |
Hb_000284_100 |
0.2643892897 |
- |
- |
PREDICTED: transcription factor GTE11 isoform X2 [Populus euphratica] |
| 12 |
Hb_000057_050 |
0.2656484728 |
- |
- |
hypothetical protein Csa_7G132920 [Cucumis sativus] |
| 13 |
Hb_002233_090 |
0.2731653344 |
- |
- |
PREDICTED: bromodomain-containing protein 3 [Jatropha curcas] |
| 14 |
Hb_001959_050 |
0.2794500385 |
- |
- |
PREDICTED: uncharacterized protein LOC103323811 isoform X1 [Prunus mume] |
| 15 |
Hb_000932_100 |
0.2843967025 |
- |
- |
- |
| 16 |
Hb_006452_090 |
0.288605903 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 5B [Jatropha curcas] |
| 17 |
Hb_003206_020 |
0.2900475997 |
- |
- |
PREDICTED: putative vacuolar protein sorting-associated protein 13A isoform X2 [Vitis vinifera] |
| 18 |
Hb_006816_170 |
0.2945777665 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump 1-like [Jatropha curcas] |
| 19 |
Hb_001912_080 |
0.2953459919 |
- |
- |
vacuolar protein sorting-associated protein, putative [Ricinus communis] |
| 20 |
Hb_000363_460 |
0.2965278868 |
- |
- |
PREDICTED: grpE protein homolog, mitochondrial isoform X1 [Jatropha curcas] |