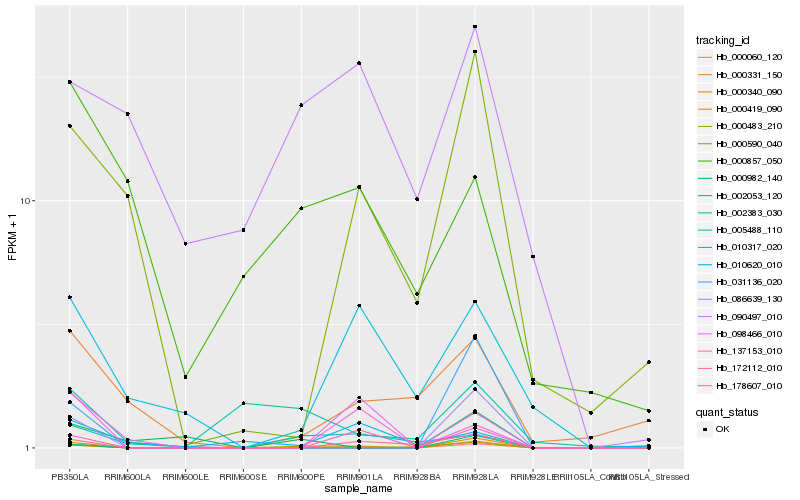
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000419_090 |
0.0 |
- |
- |
hypothetical protein JCGZ_18532 [Jatropha curcas] |
| 2 |
Hb_000060_120 |
0.3180106675 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 isoform X2 [Populus euphratica] |
| 3 |
Hb_172112_010 |
0.3298636306 |
- |
- |
hypothetical protein JCGZ_26322 [Jatropha curcas] |
| 4 |
Hb_178607_010 |
0.3444154281 |
- |
- |
PREDICTED: uncharacterized protein LOC105789535 [Gossypium raimondii] |
| 5 |
Hb_005488_110 |
0.3464810759 |
- |
- |
PREDICTED: cytochrome P450 89A2-like [Populus euphratica] |
| 6 |
Hb_086639_130 |
0.3645448457 |
- |
- |
- |
| 7 |
Hb_137153_010 |
0.3732027215 |
- |
- |
- |
| 8 |
Hb_098466_010 |
0.3802223152 |
- |
- |
PREDICTED: uncharacterized protein LOC105640954 [Jatropha curcas] |
| 9 |
Hb_000590_040 |
0.3880750372 |
- |
- |
hypothetical protein MPER_09217 [Moniliophthora perniciosa FA553] |
| 10 |
Hb_010317_020 |
0.3946610894 |
- |
- |
PREDICTED: sarcoplasmic reticulum histidine-rich calcium-binding protein [Jatropha curcas] |
| 11 |
Hb_000982_140 |
0.395013508 |
- |
- |
histidine kinase 1 plant, putative [Ricinus communis] |
| 12 |
Hb_002383_030 |
0.3970025018 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 13 |
Hb_002053_120 |
0.397201296 |
transcription factor |
TF Family: CSD |
PREDICTED: uncharacterized protein LOC105636942 [Jatropha curcas] |
| 14 |
Hb_031136_020 |
0.3983960028 |
- |
- |
- |
| 15 |
Hb_000340_090 |
0.3985857059 |
- |
- |
- |
| 16 |
Hb_000331_150 |
0.4131490034 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 17 |
Hb_000483_210 |
0.4219408098 |
- |
- |
hypothetical protein JCGZ_03497 [Jatropha curcas] |
| 18 |
Hb_090497_010 |
0.422426814 |
- |
- |
PREDICTED: Werner Syndrome-like exonuclease [Jatropha curcas] |
| 19 |
Hb_010620_010 |
0.4274342726 |
- |
- |
hypothetical protein JCGZ_22570 [Jatropha curcas] |
| 20 |
Hb_000857_050 |
0.4285686573 |
- |
- |
PREDICTED: protein YLS9 [Jatropha curcas] |