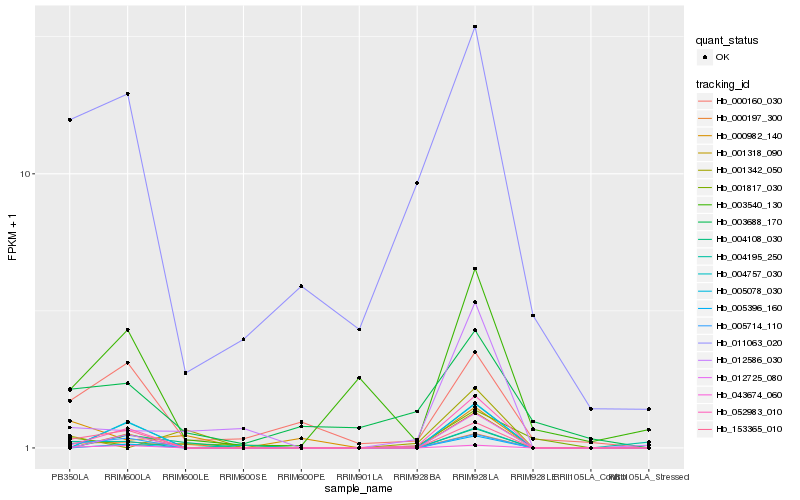
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000197_300 |
0.0 |
- |
- |
dehydrin protein [Manihot esculenta] |
| 2 |
Hb_153365_010 |
0.2700897165 |
- |
- |
PREDICTED: uncharacterized protein LOC104236681 [Nicotiana sylvestris] |
| 3 |
Hb_005714_110 |
0.2763088142 |
- |
- |
PREDICTED: probable indole-3-pyruvate monooxygenase YUCCA10 [Jatropha curcas] |
| 4 |
Hb_000982_140 |
0.2864916447 |
- |
- |
histidine kinase 1 plant, putative [Ricinus communis] |
| 5 |
Hb_043674_060 |
0.2976959669 |
- |
- |
PREDICTED: uncharacterized protein LOC104240691 [Nicotiana sylvestris] |
| 6 |
Hb_004108_030 |
0.3051294295 |
- |
- |
- |
| 7 |
Hb_052983_010 |
0.3455027214 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 8 |
Hb_012586_030 |
0.3557081565 |
- |
- |
hypothetical protein CICLE_v10014868mg [Citrus clementina] |
| 9 |
Hb_000160_030 |
0.3575574593 |
- |
- |
putative S-adenosylmethionine-dependent methyltransferase [Morus notabilis] |
| 10 |
Hb_001342_050 |
0.3698423294 |
- |
- |
- |
| 11 |
Hb_003540_130 |
0.3727717634 |
- |
- |
- |
| 12 |
Hb_011063_020 |
0.3836200661 |
- |
- |
PREDICTED: ribosomal L1 domain-containing protein 1-like [Jatropha curcas] |
| 13 |
Hb_001817_030 |
0.3910824078 |
- |
- |
PREDICTED: putative CCR4-associated factor 1 homolog 8 [Jatropha curcas] |
| 14 |
Hb_003688_170 |
0.395500857 |
- |
- |
PREDICTED: presenilin-like protein At2g29900 isoform X2 [Cucumis sativus] |
| 15 |
Hb_001318_090 |
0.3981556687 |
- |
- |
PREDICTED: zinc finger protein 598 [Jatropha curcas] |
| 16 |
Hb_012725_080 |
0.3982124266 |
- |
- |
- |
| 17 |
Hb_005396_160 |
0.4000330662 |
- |
- |
PREDICTED: clavaminate synthase-like protein At3g21360 [Jatropha curcas] |
| 18 |
Hb_004757_030 |
0.4015300912 |
- |
- |
- |
| 19 |
Hb_005078_030 |
0.4017141635 |
- |
- |
- |
| 20 |
Hb_004195_250 |
0.4047251506 |
- |
- |
hypothetical protein PRUPE_ppa015658mg [Prunus persica] |