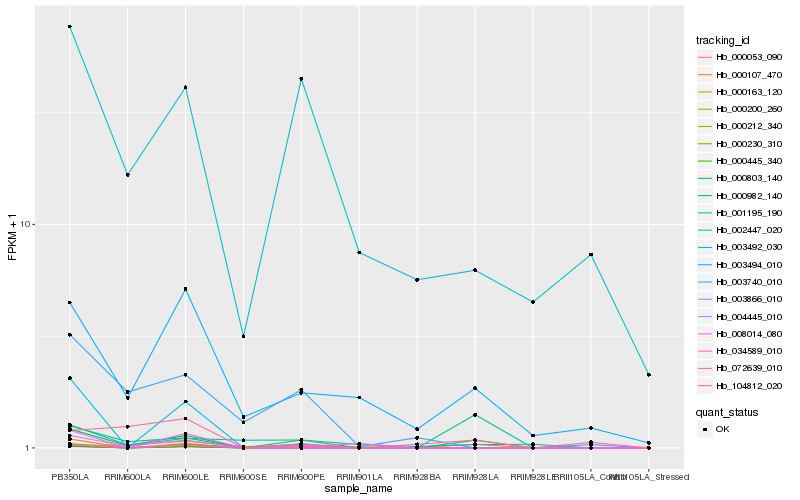
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000803_140 |
0.0 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase-like [Populus euphratica] |
| 2 |
Hb_000982_140 |
0.288722588 |
- |
- |
histidine kinase 1 plant, putative [Ricinus communis] |
| 3 |
Hb_003494_010 |
0.3144521204 |
- |
- |
hypothetical protein JCGZ_19838 [Jatropha curcas] |
| 4 |
Hb_034589_010 |
0.3612990914 |
- |
- |
hypothetical protein MIMGU_mgv1a020753mg, partial [Erythranthe guttata] |
| 5 |
Hb_002447_020 |
0.3620077847 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_104812_020 |
0.3829764955 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 7 |
Hb_000107_470 |
0.384832896 |
- |
- |
cellulose synthase, putative [Ricinus communis] |
| 8 |
Hb_003740_010 |
0.3922380135 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000230_310 |
0.3977110104 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 10 |
Hb_001195_190 |
0.4060924337 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004445_010 |
0.4073019761 |
- |
- |
kinase family protein [Populus trichocarpa] |
| 12 |
Hb_003492_030 |
0.408984599 |
- |
- |
- |
| 13 |
Hb_003866_010 |
0.4098138271 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 14 |
Hb_000445_340 |
0.4104992301 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000053_090 |
0.4108480892 |
- |
- |
PREDICTED: clavaminate synthase-like protein At3g21360 [Populus euphratica] |
| 16 |
Hb_072639_010 |
0.4108644136 |
- |
- |
JHL22C18.10 [Jatropha curcas] |
| 17 |
Hb_008014_080 |
0.4109697022 |
- |
- |
PREDICTED: uncharacterized protein LOC103848614 [Brassica rapa] |
| 18 |
Hb_000212_340 |
0.4110769847 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000163_120 |
0.4111053912 |
- |
- |
- |
| 20 |
Hb_000200_260 |
0.4114773008 |
- |
- |
PREDICTED: uncharacterized protein LOC104232531 isoform X2 [Nicotiana sylvestris] |