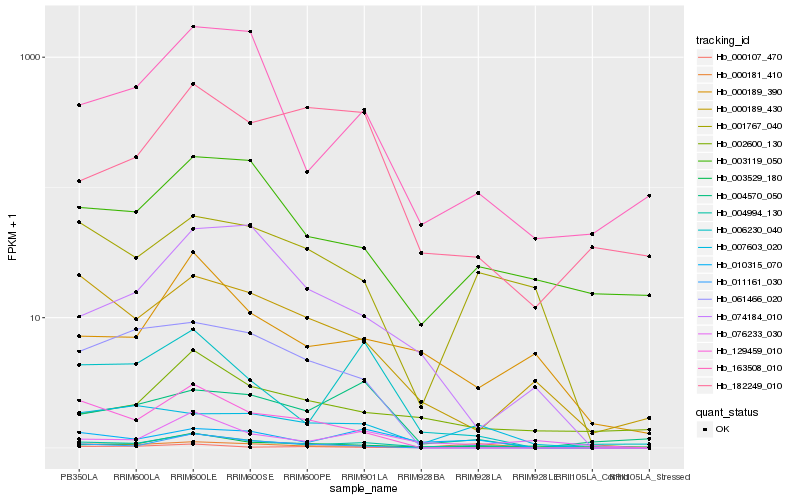
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004994_130 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-like protein M [Jatropha curcas] |
| 2 |
Hb_010315_070 |
0.1713078316 |
- |
- |
- |
| 3 |
Hb_000181_410 |
0.2190193462 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_006230_040 |
0.2206056197 |
- |
- |
kinase, putative [Ricinus communis] |
| 5 |
Hb_163508_010 |
0.2286795741 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 6 |
Hb_000189_390 |
0.2355198492 |
- |
- |
PREDICTED: pollen-specific protein SF21-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_007603_020 |
0.2377707208 |
- |
- |
PREDICTED: leucine-rich repeat receptor protein kinase MSP1-like [Jatropha curcas] |
| 8 |
Hb_003119_050 |
0.2403431192 |
- |
- |
PREDICTED: bifunctional riboflavin biosynthesis protein RIBA 1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_129459_010 |
0.2418966647 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g18840-like [Populus euphratica] |
| 10 |
Hb_076233_030 |
0.2427213521 |
- |
- |
PREDICTED: phosphoserine phosphatase, chloroplastic [Jatropha curcas] |
| 11 |
Hb_003529_180 |
0.2464501579 |
- |
- |
hypothetical protein JCGZ_00222 [Jatropha curcas] |
| 12 |
Hb_001767_040 |
0.247849606 |
- |
- |
dehydroascorbate reductase, putative [Ricinus communis] |
| 13 |
Hb_011161_030 |
0.247893549 |
- |
- |
PREDICTED: uncharacterized protein LOC105638906 [Jatropha curcas] |
| 14 |
Hb_004570_050 |
0.2484033025 |
- |
- |
kinase, putative [Ricinus communis] |
| 15 |
Hb_000107_470 |
0.249762855 |
- |
- |
cellulose synthase, putative [Ricinus communis] |
| 16 |
Hb_000189_430 |
0.2531310575 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_074184_010 |
0.2535493315 |
- |
- |
eukaryotic translation initiation factor 3 subunit, putative [Ricinus communis] |
| 18 |
Hb_002600_130 |
0.2553132403 |
- |
- |
PREDICTED: uncharacterized protein LOC105646808 [Jatropha curcas] |
| 19 |
Hb_061466_020 |
0.2585155832 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 20 |
Hb_182249_010 |
0.2618543645 |
- |
- |
- |