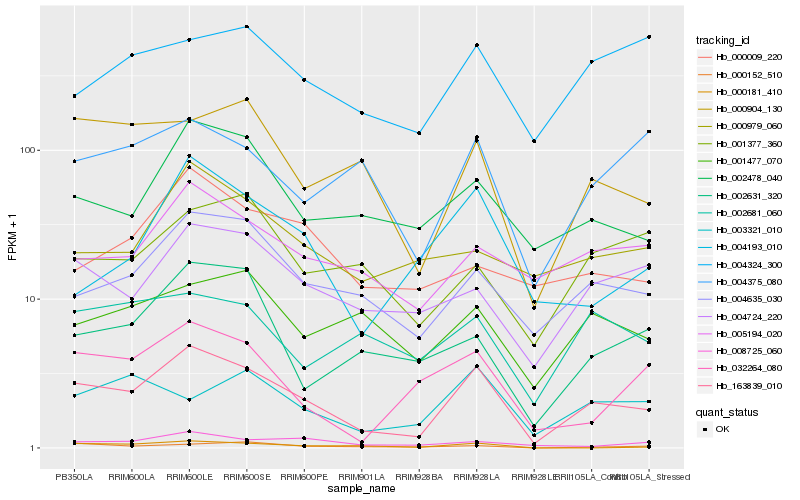
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_163839_010 |
0.0 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 2 |
Hb_004635_030 |
0.1843933975 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g57790-like [Jatropha curcas] |
| 3 |
Hb_002478_040 |
0.1963707783 |
transcription factor |
TF Family: ERF |
Dehydration-responsive element-binding protein 2C, putative [Ricinus communis] |
| 4 |
Hb_000904_130 |
0.1968618911 |
- |
- |
PREDICTED: protein CYPRO4 [Jatropha curcas] |
| 5 |
Hb_004724_220 |
0.2001881173 |
- |
- |
PREDICTED: probable calcium-binding protein CML48 [Jatropha curcas] |
| 6 |
Hb_005194_020 |
0.2090743424 |
- |
- |
PREDICTED: UPF0553 protein-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_032264_080 |
0.2129494611 |
- |
- |
PREDICTED: uncharacterized protein LOC105648216 [Jatropha curcas] |
| 8 |
Hb_004193_010 |
0.2166941333 |
- |
- |
PREDICTED: uncharacterized protein LOC105650669 [Jatropha curcas] |
| 9 |
Hb_008725_060 |
0.2188148486 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 10 |
Hb_002631_320 |
0.2192330844 |
- |
- |
hypothetical protein JCGZ_22628 [Jatropha curcas] |
| 11 |
Hb_001477_070 |
0.2197979839 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002681_060 |
0.2218571187 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |
| 13 |
Hb_004375_080 |
0.2219591688 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF073 [Jatropha curcas] |
| 14 |
Hb_003321_010 |
0.2221056794 |
- |
- |
hypothetical protein RCOM_0884420 [Ricinus communis] |
| 15 |
Hb_000152_510 |
0.2247218994 |
- |
- |
kinase, putative [Ricinus communis] |
| 16 |
Hb_000979_060 |
0.2263058646 |
- |
- |
PREDICTED: protein PLANT CADMIUM RESISTANCE 2-like [Gossypium raimondii] |
| 17 |
Hb_001377_360 |
0.2296557352 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 18 [Jatropha curcas] |
| 18 |
Hb_004324_300 |
0.2300445685 |
- |
- |
hypothetical protein PRUPE_ppa012628mg [Prunus persica] |
| 19 |
Hb_000009_220 |
0.2304026522 |
- |
- |
PREDICTED: calcium uniporter protein 2, mitochondrial-like [Jatropha curcas] |
| 20 |
Hb_000181_410 |
0.230863741 |
- |
- |
conserved hypothetical protein [Ricinus communis] |