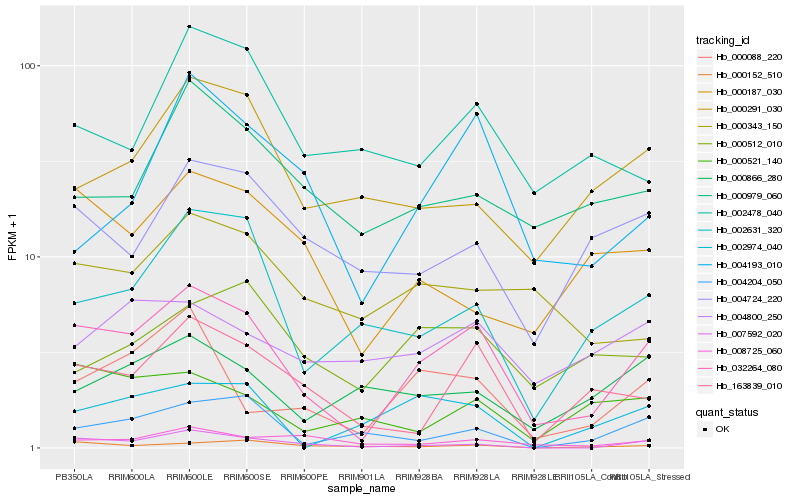
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032264_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648216 [Jatropha curcas] |
| 2 |
Hb_007592_020 |
0.2027356231 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 3 |
Hb_163839_010 |
0.2129494611 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 4 |
Hb_002631_320 |
0.2187577606 |
- |
- |
hypothetical protein JCGZ_22628 [Jatropha curcas] |
| 5 |
Hb_000088_220 |
0.2188251119 |
transcription factor |
TF Family: zf-HD |
transcription factor, putative [Ricinus communis] |
| 6 |
Hb_004724_220 |
0.2297337966 |
- |
- |
PREDICTED: probable calcium-binding protein CML48 [Jatropha curcas] |
| 7 |
Hb_000187_030 |
0.2337713188 |
- |
- |
PREDICTED: cyclin-D4-1-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_004193_010 |
0.235627646 |
- |
- |
PREDICTED: uncharacterized protein LOC105650669 [Jatropha curcas] |
| 9 |
Hb_008725_060 |
0.2356673993 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 10 |
Hb_000152_510 |
0.2393439855 |
- |
- |
kinase, putative [Ricinus communis] |
| 11 |
Hb_000291_030 |
0.240869243 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1-like [Eucalyptus grandis] |
| 12 |
Hb_000979_060 |
0.241641689 |
- |
- |
PREDICTED: protein PLANT CADMIUM RESISTANCE 2-like [Gossypium raimondii] |
| 13 |
Hb_004800_250 |
0.2430794141 |
- |
- |
PREDICTED: protein N-lysine methyltransferase METTL21A [Jatropha curcas] |
| 14 |
Hb_002478_040 |
0.2453914941 |
transcription factor |
TF Family: ERF |
Dehydration-responsive element-binding protein 2C, putative [Ricinus communis] |
| 15 |
Hb_000866_280 |
0.2455987642 |
- |
- |
PREDICTED: seven transmembrane domain-containing tyrosine-protein kinase 1-like [Jatropha curcas] |
| 16 |
Hb_002974_040 |
0.2456558387 |
- |
- |
GRF zinc finger containing protein [Medicago truncatula] |
| 17 |
Hb_000521_140 |
0.2457610653 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Malus domestica] |
| 18 |
Hb_000343_150 |
0.245807957 |
- |
- |
PREDICTED: MAR-binding filament-like protein 1-1 [Jatropha curcas] |
| 19 |
Hb_000512_010 |
0.2463231318 |
- |
- |
PREDICTED: uncharacterized protein LOC105650728 [Jatropha curcas] |
| 20 |
Hb_004204_050 |
0.2464124506 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |