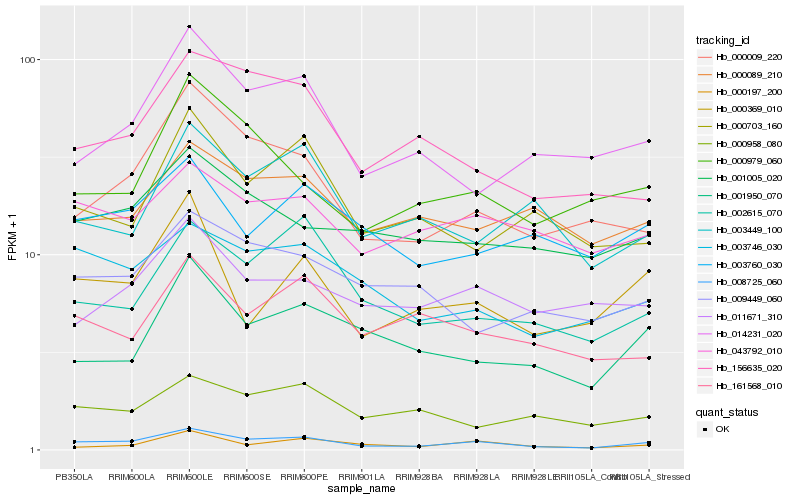
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008725_060 |
0.0 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 2 |
Hb_000009_220 |
0.1313105445 |
- |
- |
PREDICTED: calcium uniporter protein 2, mitochondrial-like [Jatropha curcas] |
| 3 |
Hb_003746_030 |
0.1470836005 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 2 [Jatropha curcas] |
| 4 |
Hb_000369_010 |
0.1471888852 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002615_070 |
0.1475340259 |
- |
- |
PREDICTED: uncharacterized protein LOC105628992 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001950_070 |
0.1525205765 |
- |
- |
cyclin d, putative [Ricinus communis] |
| 7 |
Hb_014231_020 |
0.1534469427 |
- |
- |
PREDICTED: ras-related protein Rab11D [Jatropha curcas] |
| 8 |
Hb_000197_200 |
0.1541356467 |
- |
- |
Sec14 cytosolic factor, putative [Ricinus communis] |
| 9 |
Hb_000979_060 |
0.1547156504 |
- |
- |
PREDICTED: protein PLANT CADMIUM RESISTANCE 2-like [Gossypium raimondii] |
| 10 |
Hb_156635_020 |
0.1549752739 |
- |
- |
PREDICTED: L-lactate dehydrogenase B [Jatropha curcas] |
| 11 |
Hb_043792_010 |
0.1558551581 |
- |
- |
PREDICTED: uncharacterized protein LOC105645113 isoform X2 [Jatropha curcas] |
| 12 |
Hb_001005_020 |
0.1577629193 |
- |
- |
PREDICTED: uncharacterized protein LOC105630212 [Jatropha curcas] |
| 13 |
Hb_161568_010 |
0.1622127902 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 14 |
Hb_000089_210 |
0.16253516 |
- |
- |
unknown [Medicago truncatula] |
| 15 |
Hb_000703_160 |
0.1626370778 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_003760_030 |
0.1628649231 |
- |
- |
PREDICTED: novel plant SNARE 13 [Jatropha curcas] |
| 17 |
Hb_009449_060 |
0.1631947665 |
- |
- |
PREDICTED: ubiquitin-like-specific protease 1D isoform X2 [Jatropha curcas] |
| 18 |
Hb_003449_100 |
0.1633312499 |
- |
- |
PREDICTED: protein YIPF6 homolog [Jatropha curcas] |
| 19 |
Hb_000958_080 |
0.1637759689 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_011671_310 |
0.163840385 |
- |
- |
PREDICTED: polycomb group protein EMBRYONIC FLOWER 2 [Jatropha curcas] |