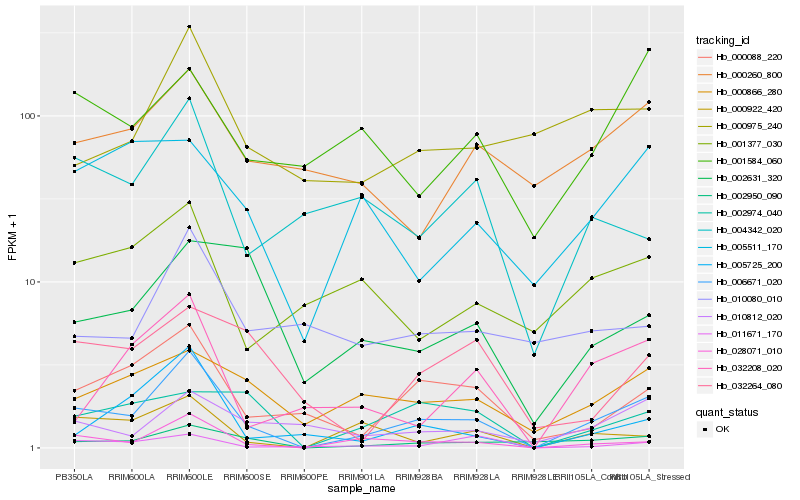
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006671_020 |
0.0 |
- |
- |
hypothetical protein CISIN_1g0407181mg, partial [Citrus sinensis] |
| 2 |
Hb_028071_010 |
0.1927030757 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |
| 3 |
Hb_002950_090 |
0.233798607 |
- |
- |
- |
| 4 |
Hb_000088_220 |
0.2414251435 |
transcription factor |
TF Family: zf-HD |
transcription factor, putative [Ricinus communis] |
| 5 |
Hb_000922_420 |
0.2643594247 |
- |
- |
glutaredoxin-1, grx1, putative [Ricinus communis] |
| 6 |
Hb_005725_200 |
0.2645744941 |
transcription factor |
TF Family: GRF |
hypothetical protein JCGZ_15989 [Jatropha curcas] |
| 7 |
Hb_000866_280 |
0.2665524176 |
- |
- |
PREDICTED: seven transmembrane domain-containing tyrosine-protein kinase 1-like [Jatropha curcas] |
| 8 |
Hb_000975_240 |
0.2732635177 |
- |
- |
hypothetical protein JCGZ_22006 [Jatropha curcas] |
| 9 |
Hb_010812_020 |
0.2738040455 |
transcription factor |
TF Family: IWS1 |
transcription elongation factor s-II, putative [Ricinus communis] |
| 10 |
Hb_032208_020 |
0.2753468076 |
- |
- |
hypothetical protein JCGZ_01894 [Jatropha curcas] |
| 11 |
Hb_004342_020 |
0.2794337033 |
- |
- |
PREDICTED: serine/threonine-protein kinase Aurora-1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_011671_170 |
0.2832239304 |
- |
- |
PREDICTED: protein DYAD [Jatropha curcas] |
| 13 |
Hb_002974_040 |
0.2850721779 |
- |
- |
GRF zinc finger containing protein [Medicago truncatula] |
| 14 |
Hb_000260_800 |
0.2879664001 |
- |
- |
PREDICTED: probable NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 12 [Populus euphratica] |
| 15 |
Hb_005511_170 |
0.2881085309 |
- |
- |
hypothetical protein CISIN_1g030718mg [Citrus sinensis] |
| 16 |
Hb_001377_030 |
0.2906241826 |
- |
- |
PREDICTED: histone H3-like centromeric protein HTR12 [Jatropha curcas] |
| 17 |
Hb_010080_010 |
0.2931859919 |
- |
- |
PREDICTED: uncharacterized protein LOC105635247 [Jatropha curcas] |
| 18 |
Hb_002631_320 |
0.2946033411 |
- |
- |
hypothetical protein JCGZ_22628 [Jatropha curcas] |
| 19 |
Hb_001584_060 |
0.2955046294 |
- |
- |
PREDICTED: transcription factor bHLH149 [Jatropha curcas] |
| 20 |
Hb_032264_080 |
0.2994178409 |
- |
- |
PREDICTED: uncharacterized protein LOC105648216 [Jatropha curcas] |