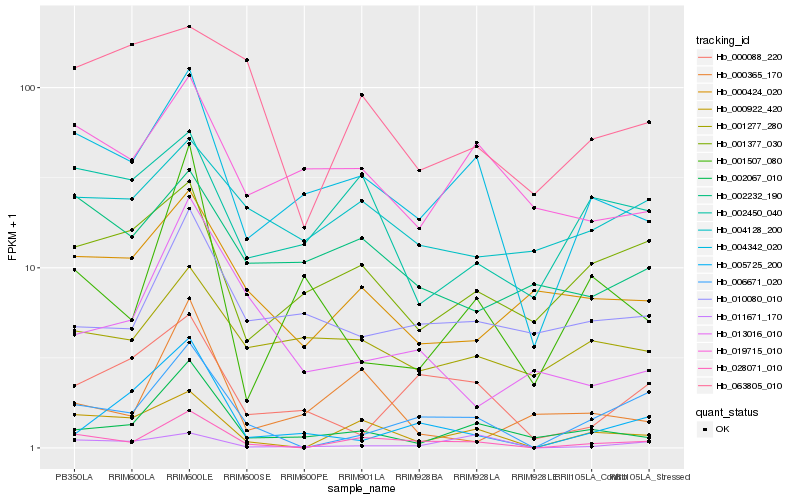
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028071_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |
| 2 |
Hb_000922_420 |
0.1878667875 |
- |
- |
glutaredoxin-1, grx1, putative [Ricinus communis] |
| 3 |
Hb_006671_020 |
0.1927030757 |
- |
- |
hypothetical protein CISIN_1g0407181mg, partial [Citrus sinensis] |
| 4 |
Hb_004342_020 |
0.2356958654 |
- |
- |
PREDICTED: serine/threonine-protein kinase Aurora-1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_002067_010 |
0.2719857163 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000424_020 |
0.2947944134 |
- |
- |
Uncharacterized protein TCM_037386 [Theobroma cacao] |
| 7 |
Hb_013016_010 |
0.3025900422 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002450_040 |
0.3041052302 |
- |
- |
PREDICTED: DNA-directed RNA polymerases II and V subunit 6B [Vitis vinifera] |
| 9 |
Hb_000365_170 |
0.3060667934 |
- |
- |
PREDICTED: 50S ribosomal protein L17, chloroplastic isoform X3 [Jatropha curcas] |
| 10 |
Hb_001377_030 |
0.3075883365 |
- |
- |
PREDICTED: histone H3-like centromeric protein HTR12 [Jatropha curcas] |
| 11 |
Hb_010080_010 |
0.3078635118 |
- |
- |
PREDICTED: uncharacterized protein LOC105635247 [Jatropha curcas] |
| 12 |
Hb_002232_190 |
0.308364265 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_019715_010 |
0.3088051596 |
- |
- |
carbon catabolite repressor protein, putative [Ricinus communis] |
| 14 |
Hb_000088_220 |
0.3111563579 |
transcription factor |
TF Family: zf-HD |
transcription factor, putative [Ricinus communis] |
| 15 |
Hb_011671_170 |
0.3148670045 |
- |
- |
PREDICTED: protein DYAD [Jatropha curcas] |
| 16 |
Hb_001277_280 |
0.3155922041 |
- |
- |
PREDICTED: protein argonaute 10 [Jatropha curcas] |
| 17 |
Hb_001507_080 |
0.3159167142 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin S13-7 [Jatropha curcas] |
| 18 |
Hb_005725_200 |
0.3187810231 |
transcription factor |
TF Family: GRF |
hypothetical protein JCGZ_15989 [Jatropha curcas] |
| 19 |
Hb_063805_010 |
0.3191340602 |
- |
- |
- |
| 20 |
Hb_004128_200 |
0.3194135305 |
- |
- |
PREDICTED: uncharacterized protein LOC105642137 [Jatropha curcas] |