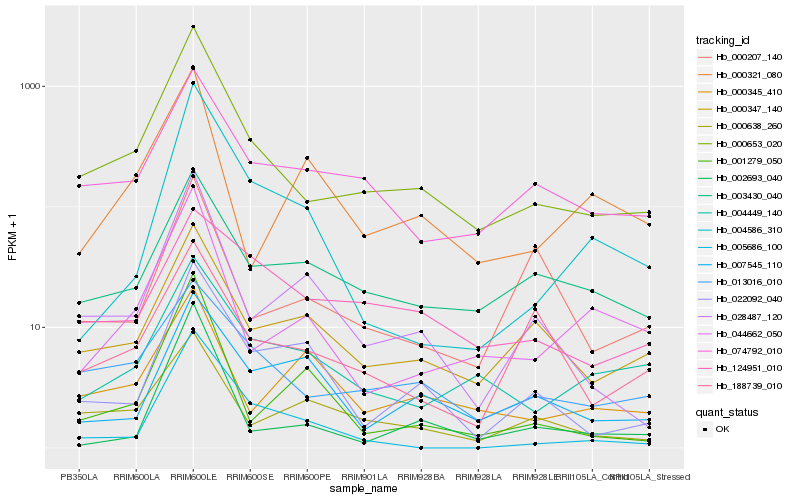
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000653_020 |
0.0 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15-like [Jatropha curcas] |
| 2 |
Hb_013016_010 |
0.1585170024 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_074792_010 |
0.1809390653 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast [Gossypium raimondii] |
| 4 |
Hb_000347_140 |
0.1868249586 |
- |
- |
PREDICTED: protein FLUORESCENT IN BLUE LIGHT, chloroplastic isoform X2 [Jatropha curcas] |
| 5 |
Hb_028487_120 |
0.1873739991 |
- |
- |
PREDICTED: HMG-Y-related protein B-like [Jatropha curcas] |
| 6 |
Hb_044662_050 |
0.1919532794 |
- |
- |
hypothetical protein POPTR_0002s08440g [Populus trichocarpa] |
| 7 |
Hb_004449_140 |
0.1940066273 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor GAMYB-like [Jatropha curcas] |
| 8 |
Hb_000638_260 |
0.1982281583 |
- |
- |
hypothetical protein JCGZ_06730 [Jatropha curcas] |
| 9 |
Hb_003430_040 |
0.1987124653 |
- |
- |
spermidine synthase 1, putative [Ricinus communis] |
| 10 |
Hb_022092_040 |
0.2132959455 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 1 [Jatropha curcas] |
| 11 |
Hb_188739_010 |
0.2225096382 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 12 |
Hb_005686_100 |
0.2238692614 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004586_310 |
0.2238856298 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002693_040 |
0.2245943434 |
- |
- |
hypersensitive-induced response protein [Carica papaya] |
| 15 |
Hb_001279_050 |
0.2285565294 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 16 |
Hb_000321_080 |
0.2292699558 |
- |
- |
histone H4 [Zea mays] |
| 17 |
Hb_124951_010 |
0.2340498978 |
- |
- |
chitinase-like protein [Hevea brasiliensis] |
| 18 |
Hb_000345_410 |
0.2350661378 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 [Jatropha curcas] |
| 19 |
Hb_000207_140 |
0.2369279767 |
- |
- |
hypothetical protein POPTR_0019s11310g [Populus trichocarpa] |
| 20 |
Hb_007545_110 |
0.2379459169 |
- |
- |
catalytic, putative [Ricinus communis] |