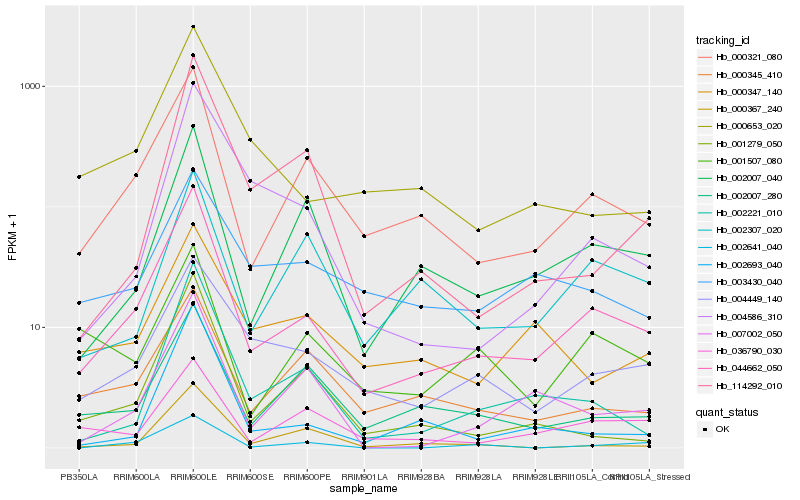
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_044662_050 |
0.0 |
- |
- |
hypothetical protein POPTR_0002s08440g [Populus trichocarpa] |
| 2 |
Hb_000321_080 |
0.133258101 |
- |
- |
histone H4 [Zea mays] |
| 3 |
Hb_002007_040 |
0.186043515 |
- |
- |
unnamed protein product [Coffea canephora] |
| 4 |
Hb_002221_010 |
0.1871820376 |
- |
- |
PREDICTED: adenylyl-sulfate kinase 3-like [Jatropha curcas] |
| 5 |
Hb_004449_140 |
0.1889266459 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor GAMYB-like [Jatropha curcas] |
| 6 |
Hb_000653_020 |
0.1919532794 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15-like [Jatropha curcas] |
| 7 |
Hb_002007_280 |
0.1961830364 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 [Jatropha curcas] |
| 8 |
Hb_007002_050 |
0.2002726107 |
- |
- |
PREDICTED: uncharacterized protein LOC105642607 [Jatropha curcas] |
| 9 |
Hb_002693_040 |
0.2042464696 |
- |
- |
hypersensitive-induced response protein [Carica papaya] |
| 10 |
Hb_000345_410 |
0.2052244633 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 [Jatropha curcas] |
| 11 |
Hb_001279_050 |
0.2053935251 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 12 |
Hb_036790_030 |
0.2131829082 |
- |
- |
PREDICTED: UPF0544 protein C5orf45 homolog [Jatropha curcas] |
| 13 |
Hb_004586_310 |
0.2149342689 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002641_040 |
0.217585056 |
- |
- |
class III peroxidase [Populus trichocarpa] |
| 15 |
Hb_000347_140 |
0.2225015574 |
- |
- |
PREDICTED: protein FLUORESCENT IN BLUE LIGHT, chloroplastic isoform X2 [Jatropha curcas] |
| 16 |
Hb_001507_080 |
0.2241778084 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin S13-7 [Jatropha curcas] |
| 17 |
Hb_003430_040 |
0.2247712839 |
- |
- |
spermidine synthase 1, putative [Ricinus communis] |
| 18 |
Hb_002307_020 |
0.2249408495 |
- |
- |
PREDICTED: histone H1-like [Jatropha curcas] |
| 19 |
Hb_114292_010 |
0.2270934727 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000367_240 |
0.2285249168 |
- |
- |
hypothetical protein JCGZ_11682 [Jatropha curcas] |