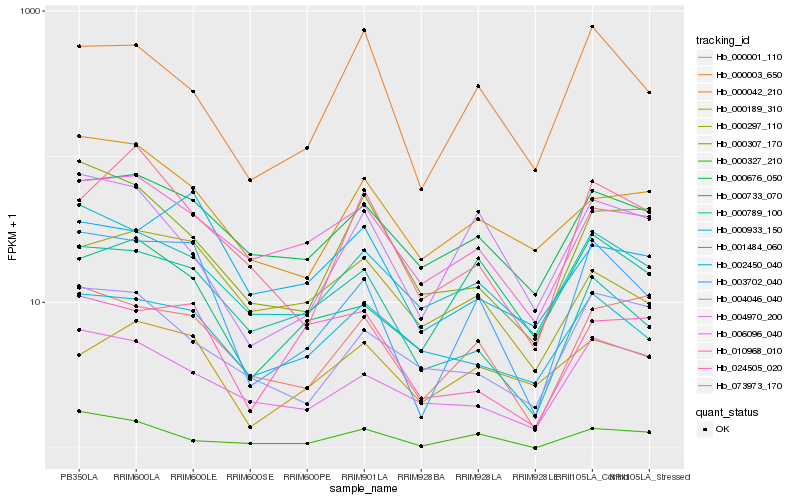
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003702_040 |
0.0 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000001_110 |
0.1785451476 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000789_100 |
0.1803255563 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 13-like [Jatropha curcas] |
| 4 |
Hb_000933_150 |
0.1833357908 |
- |
- |
PREDICTED: mitotic checkpoint protein BUB3.3 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000676_050 |
0.1919894908 |
- |
- |
PREDICTED: thioredoxin domain-containing protein PLP3B-like [Gossypium raimondii] |
| 6 |
Hb_000003_650 |
0.1967050231 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 7 |
Hb_006096_040 |
0.1975802857 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 8 [Jatropha curcas] |
| 8 |
Hb_004970_200 |
0.1982921199 |
- |
- |
serine/threonine-protein kinase, putative [Ricinus communis] |
| 9 |
Hb_000297_110 |
0.1997988453 |
- |
- |
PREDICTED: uncharacterized protein LOC105634718 [Jatropha curcas] |
| 10 |
Hb_010968_010 |
0.2040110678 |
- |
- |
Golgi snare 12 isoform 1 [Theobroma cacao] |
| 11 |
Hb_000733_070 |
0.2059597439 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 5 [Jatropha curcas] |
| 12 |
Hb_001484_060 |
0.2085027224 |
- |
- |
PREDICTED: uncharacterized protein LOC105641630 [Jatropha curcas] |
| 13 |
Hb_000042_210 |
0.2108908527 |
- |
- |
PREDICTED: uncharacterized protein LOC105632808 [Jatropha curcas] |
| 14 |
Hb_002450_040 |
0.2110111428 |
- |
- |
PREDICTED: DNA-directed RNA polymerases II and V subunit 6B [Vitis vinifera] |
| 15 |
Hb_000307_170 |
0.2142197827 |
- |
- |
PREDICTED: uncharacterized protein LOC105638528 [Jatropha curcas] |
| 16 |
Hb_073973_170 |
0.2147687536 |
- |
- |
PREDICTED: uncharacterized protein LOC105638918 [Jatropha curcas] |
| 17 |
Hb_000189_310 |
0.2149117181 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_024505_020 |
0.2151511913 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g80640 [Jatropha curcas] |
| 19 |
Hb_000327_210 |
0.2153962763 |
- |
- |
- |
| 20 |
Hb_004046_040 |
0.2169791952 |
transcription factor |
TF Family: GRF |
PREDICTED: growth-regulating factor 4-like [Jatropha curcas] |