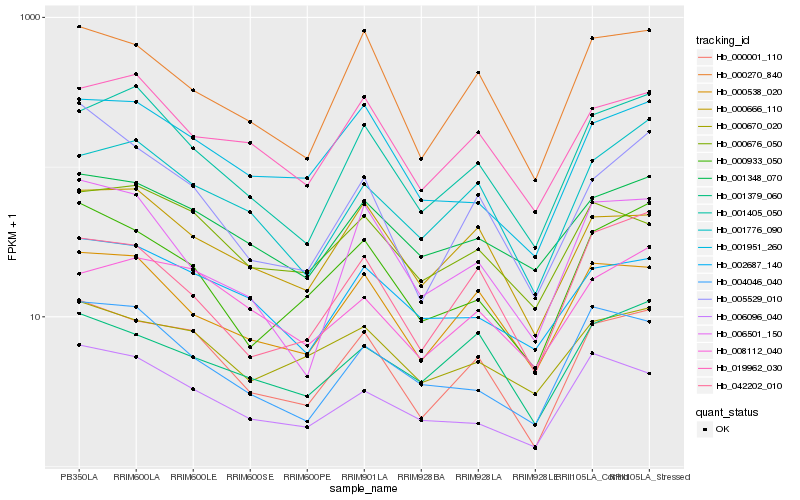
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000001_110 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000270_840 |
0.1086878652 |
- |
- |
PREDICTED: probable 26S proteasome complex subunit sem1-2 [Nicotiana tomentosiformis] |
| 3 |
Hb_000933_050 |
0.1143446392 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 4 |
Hb_000538_020 |
0.1196134031 |
- |
- |
PREDICTED: autophagy protein 5 [Jatropha curcas] |
| 5 |
Hb_001405_050 |
0.1220672502 |
- |
- |
PREDICTED: uncharacterized protein LOC100500641 [Glycine max] |
| 6 |
Hb_005529_010 |
0.1243177152 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000666_110 |
0.1263338317 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 11 [Jatropha curcas] |
| 8 |
Hb_001951_260 |
0.1308780466 |
- |
- |
60S ribosomal protein L32, putative [Ricinus communis] |
| 9 |
Hb_042202_010 |
0.1324630314 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 2 [Cucumis melo] |
| 10 |
Hb_001348_070 |
0.1331439704 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 11 |
Hb_000670_020 |
0.1333671041 |
- |
- |
PREDICTED: chromatin modification-related protein EAF3-like [Jatropha curcas] |
| 12 |
Hb_001379_060 |
0.1342207822 |
- |
- |
PREDICTED: uncharacterized protein LOC105650815 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001776_090 |
0.1365518793 |
- |
- |
mitochondrial import inner membrane translocase subunit Tim9 [Jatropha curcas] |
| 14 |
Hb_006096_040 |
0.1365792394 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 8 [Jatropha curcas] |
| 15 |
Hb_019962_030 |
0.1384060083 |
- |
- |
PREDICTED: uncharacterized protein LOC105650622 [Jatropha curcas] |
| 16 |
Hb_008112_040 |
0.1390720757 |
- |
- |
PREDICTED: uncharacterized protein LOC105637336 [Jatropha curcas] |
| 17 |
Hb_002687_140 |
0.1401109871 |
- |
- |
hypothetical protein JCGZ_24463 [Jatropha curcas] |
| 18 |
Hb_006501_150 |
0.1402216246 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000676_050 |
0.1415638582 |
- |
- |
PREDICTED: thioredoxin domain-containing protein PLP3B-like [Gossypium raimondii] |
| 20 |
Hb_004046_040 |
0.1415880637 |
transcription factor |
TF Family: GRF |
PREDICTED: growth-regulating factor 4-like [Jatropha curcas] |