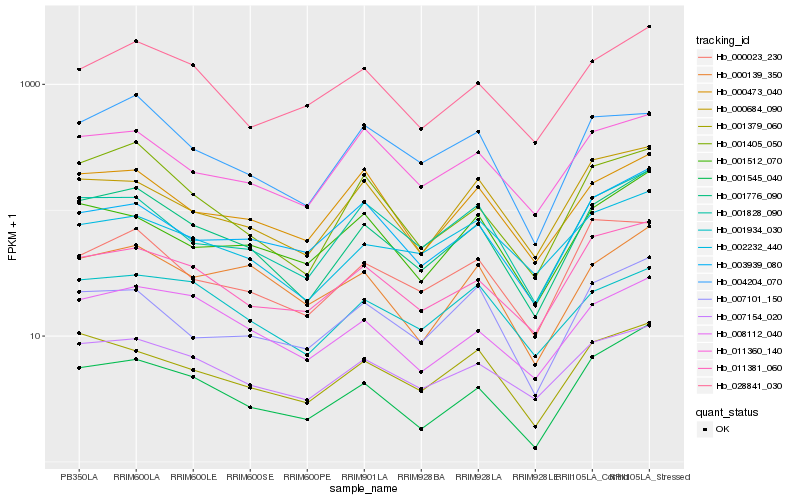
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001776_090 |
0.0 |
- |
- |
mitochondrial import inner membrane translocase subunit Tim9 [Jatropha curcas] |
| 2 |
Hb_001405_050 |
0.0900868238 |
- |
- |
PREDICTED: uncharacterized protein LOC100500641 [Glycine max] |
| 3 |
Hb_001545_040 |
0.0905000678 |
- |
- |
PREDICTED: ras-related protein Rab7-like [Malus domestica] |
| 4 |
Hb_007154_020 |
0.0923336685 |
- |
- |
- |
| 5 |
Hb_001828_090 |
0.0945617205 |
- |
- |
prohibitin, putative [Ricinus communis] |
| 6 |
Hb_011381_060 |
0.0950409555 |
- |
- |
PREDICTED: ADP-ribosylation factor 3 [Jatropha curcas] |
| 7 |
Hb_001379_060 |
0.0969941777 |
- |
- |
PREDICTED: uncharacterized protein LOC105650815 isoform X1 [Jatropha curcas] |
| 8 |
Hb_008112_040 |
0.1002128973 |
- |
- |
PREDICTED: uncharacterized protein LOC105637336 [Jatropha curcas] |
| 9 |
Hb_028841_030 |
0.1069801867 |
- |
- |
- |
| 10 |
Hb_000473_040 |
0.1109787093 |
transcription factor |
TF Family: Coactivator p15 |
PREDICTED: RNA polymerase II transcriptional coactivator KIWI-like [Jatropha curcas] |
| 11 |
Hb_001512_070 |
0.1116485982 |
- |
- |
PREDICTED: ras-related protein RABB1b [Malus domestica] |
| 12 |
Hb_000139_350 |
0.1117646675 |
- |
- |
AtGCP3 interacting protein 1 [Theobroma cacao] |
| 13 |
Hb_007101_150 |
0.1123339591 |
- |
- |
hypothetical protein CICLE_v10033984mg [Citrus clementina] |
| 14 |
Hb_000023_230 |
0.1134139119 |
- |
- |
PREDICTED: uncharacterized protein LOC105637526 [Jatropha curcas] |
| 15 |
Hb_003939_080 |
0.1137502985 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000684_090 |
0.114766906 |
transcription factor |
TF Family: S1Fa-like |
- |
| 17 |
Hb_001934_030 |
0.1149630073 |
transcription factor |
TF Family: HSF |
Heat shock factor protein, putative [Ricinus communis] |
| 18 |
Hb_011360_140 |
0.1151524917 |
- |
- |
40S ribosomal protein S19, putative [Ricinus communis] |
| 19 |
Hb_002232_440 |
0.1161061316 |
- |
- |
PREDICTED: uncharacterized protein LOC105130257 [Populus euphratica] |
| 20 |
Hb_004204_070 |
0.1164104309 |
- |
- |
PREDICTED: 40S ribosomal protein S19-3 [Jatropha curcas] |