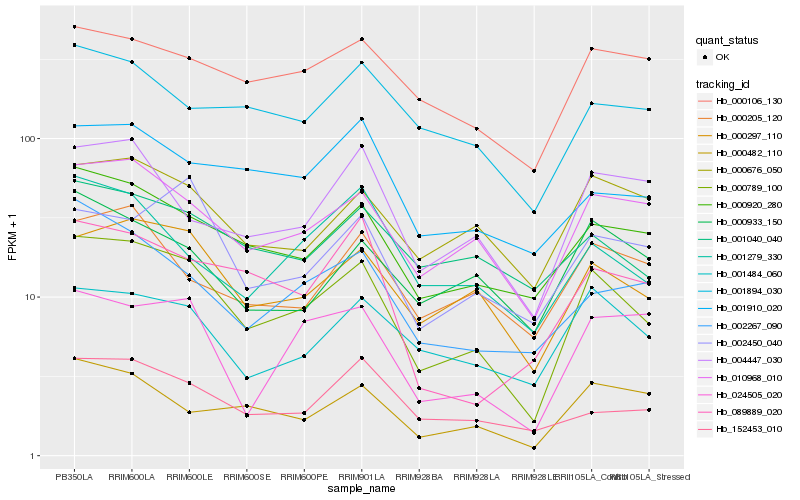
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000789_100 |
0.0 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 13-like [Jatropha curcas] |
| 2 |
Hb_000297_110 |
0.1125055389 |
- |
- |
PREDICTED: uncharacterized protein LOC105634718 [Jatropha curcas] |
| 3 |
Hb_010968_010 |
0.1218229818 |
- |
- |
Golgi snare 12 isoform 1 [Theobroma cacao] |
| 4 |
Hb_000920_280 |
0.123515584 |
- |
- |
Protein SENSITIVITY TO RED LIGHT REDUCED, putative [Ricinus communis] |
| 5 |
Hb_001484_060 |
0.1451752537 |
- |
- |
PREDICTED: uncharacterized protein LOC105641630 [Jatropha curcas] |
| 6 |
Hb_000676_050 |
0.1459568351 |
- |
- |
PREDICTED: thioredoxin domain-containing protein PLP3B-like [Gossypium raimondii] |
| 7 |
Hb_000482_110 |
0.1515088083 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 8 |
Hb_152453_010 |
0.1524575045 |
- |
- |
PREDICTED: putative transferase CAF17 homolog, mitochondrial [Jatropha curcas] |
| 9 |
Hb_001040_040 |
0.1525002655 |
- |
- |
Uncharacterized protein TCM_029905 [Theobroma cacao] |
| 10 |
Hb_002267_090 |
0.1531509208 |
- |
- |
Cytochrome c oxidase assembly protein COX19, putative [Ricinus communis] |
| 11 |
Hb_001279_330 |
0.1555704613 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 12 |
Hb_000933_150 |
0.1556126288 |
- |
- |
PREDICTED: mitotic checkpoint protein BUB3.3 isoform X1 [Jatropha curcas] |
| 13 |
Hb_004447_030 |
0.1562454618 |
- |
- |
PREDICTED: glucose-induced degradation protein 4 homolog [Jatropha curcas] |
| 14 |
Hb_024505_020 |
0.1571958067 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g80640 [Jatropha curcas] |
| 15 |
Hb_000106_130 |
0.1574711054 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_001910_020 |
0.1575945978 |
- |
- |
PREDICTED: ubiquitin-associated domain-containing protein 2 [Jatropha curcas] |
| 17 |
Hb_000205_120 |
0.1582028205 |
- |
- |
PREDICTED: casein kinase I-like isoform X3 [Jatropha curcas] |
| 18 |
Hb_001894_030 |
0.158755719 |
- |
- |
Nucleic acid-binding, OB-fold-like protein [Theobroma cacao] |
| 19 |
Hb_089889_020 |
0.1589149172 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein At2g36080-like [Jatropha curcas] |
| 20 |
Hb_002450_040 |
0.1604022495 |
- |
- |
PREDICTED: DNA-directed RNA polymerases II and V subunit 6B [Vitis vinifera] |