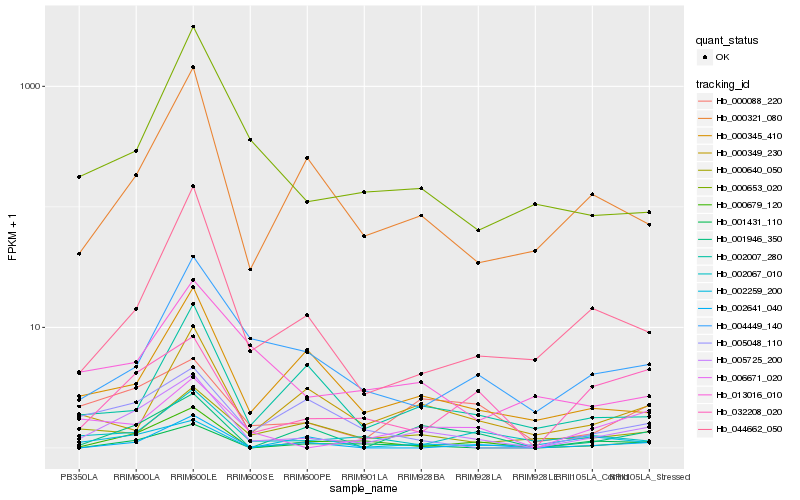
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005725_200 |
0.0 |
transcription factor |
TF Family: GRF |
hypothetical protein JCGZ_15989 [Jatropha curcas] |
| 2 |
Hb_000088_220 |
0.2266709765 |
transcription factor |
TF Family: zf-HD |
transcription factor, putative [Ricinus communis] |
| 3 |
Hb_000679_120 |
0.2289432006 |
- |
- |
hypothetical protein POPTR_0018s01610g [Populus trichocarpa] |
| 4 |
Hb_000321_080 |
0.2429992471 |
- |
- |
histone H4 [Zea mays] |
| 5 |
Hb_001946_350 |
0.2464864155 |
- |
- |
PREDICTED: uncharacterized protein LOC105639338 [Jatropha curcas] |
| 6 |
Hb_032208_020 |
0.2507235081 |
- |
- |
hypothetical protein JCGZ_01894 [Jatropha curcas] |
| 7 |
Hb_044662_050 |
0.2551860452 |
- |
- |
hypothetical protein POPTR_0002s08440g [Populus trichocarpa] |
| 8 |
Hb_006671_020 |
0.2645744941 |
- |
- |
hypothetical protein CISIN_1g0407181mg, partial [Citrus sinensis] |
| 9 |
Hb_000345_410 |
0.2654819045 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 [Jatropha curcas] |
| 10 |
Hb_004449_140 |
0.26873356 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor GAMYB-like [Jatropha curcas] |
| 11 |
Hb_000653_020 |
0.2746532496 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15-like [Jatropha curcas] |
| 12 |
Hb_013016_010 |
0.2748757999 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002067_010 |
0.276130492 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000640_050 |
0.2790929972 |
- |
- |
DNA replication licensing factor MCM7, putative [Ricinus communis] |
| 15 |
Hb_001431_110 |
0.2826901646 |
- |
- |
glycerol-3-phosphate transporter family protein [Populus trichocarpa] |
| 16 |
Hb_000349_230 |
0.2835559055 |
- |
- |
PREDICTED: protein TPX2 [Jatropha curcas] |
| 17 |
Hb_002007_280 |
0.2841397312 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 [Jatropha curcas] |
| 18 |
Hb_005048_110 |
0.2874424178 |
- |
- |
- |
| 19 |
Hb_002641_040 |
0.2880397626 |
- |
- |
class III peroxidase [Populus trichocarpa] |
| 20 |
Hb_002259_200 |
0.2885545357 |
- |
- |
conserved hypothetical protein [Ricinus communis] |