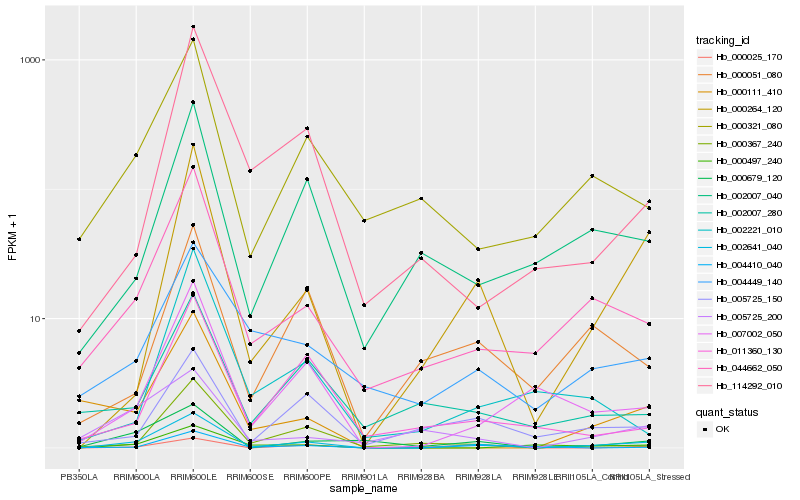
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002641_040 |
0.0 |
- |
- |
class III peroxidase [Populus trichocarpa] |
| 2 |
Hb_044662_050 |
0.217585056 |
- |
- |
hypothetical protein POPTR_0002s08440g [Populus trichocarpa] |
| 3 |
Hb_000264_120 |
0.2205971818 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 4 |
Hb_007002_050 |
0.2458774862 |
- |
- |
PREDICTED: uncharacterized protein LOC105642607 [Jatropha curcas] |
| 5 |
Hb_000679_120 |
0.2536865848 |
- |
- |
hypothetical protein POPTR_0018s01610g [Populus trichocarpa] |
| 6 |
Hb_000321_080 |
0.2591750828 |
- |
- |
histone H4 [Zea mays] |
| 7 |
Hb_002007_040 |
0.2704784572 |
- |
- |
unnamed protein product [Coffea canephora] |
| 8 |
Hb_000367_240 |
0.2713260276 |
- |
- |
hypothetical protein JCGZ_11682 [Jatropha curcas] |
| 9 |
Hb_000497_240 |
0.2733021285 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 10 |
Hb_000025_170 |
0.2763553075 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000111_410 |
0.2775436994 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_005725_150 |
0.2784776056 |
- |
- |
PREDICTED: kinesin-related protein 11 isoform X1 [Jatropha curcas] |
| 13 |
Hb_004449_140 |
0.2789019606 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor GAMYB-like [Jatropha curcas] |
| 14 |
Hb_004410_040 |
0.2812796674 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 15 |
Hb_011360_130 |
0.2839395865 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000051_080 |
0.2841177483 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase CMT3 [Jatropha curcas] |
| 17 |
Hb_114292_010 |
0.2845451754 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005725_200 |
0.2880397626 |
transcription factor |
TF Family: GRF |
hypothetical protein JCGZ_15989 [Jatropha curcas] |
| 19 |
Hb_002007_280 |
0.2891804976 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 [Jatropha curcas] |
| 20 |
Hb_002221_010 |
0.2950309303 |
- |
- |
PREDICTED: adenylyl-sulfate kinase 3-like [Jatropha curcas] |