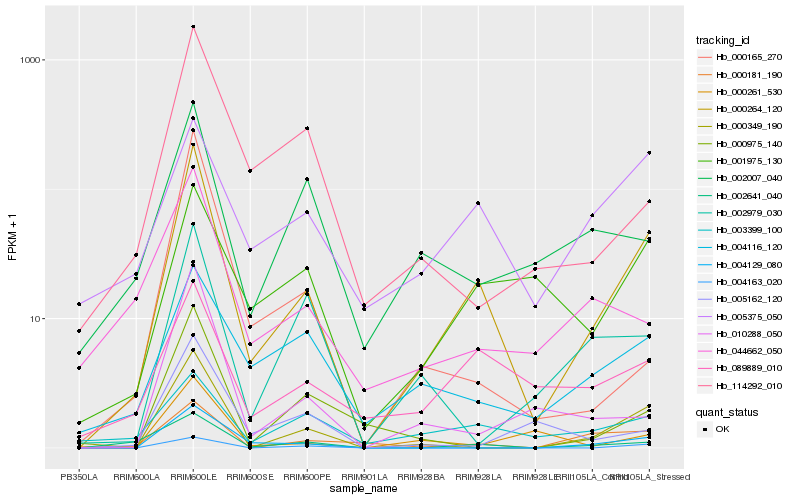
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000264_120 |
0.0 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 2 |
Hb_000349_190 |
0.217093314 |
transcription factor |
TF Family: GRF |
hypothetical protein POPTR_0001s08310g [Populus trichocarpa] |
| 3 |
Hb_002641_040 |
0.2205971818 |
- |
- |
class III peroxidase [Populus trichocarpa] |
| 4 |
Hb_004129_080 |
0.2317110521 |
- |
- |
PREDICTED: putative DNA-binding protein ESCAROLA [Prunus mume] |
| 5 |
Hb_000181_190 |
0.2618130052 |
- |
- |
PREDICTED: probable protein phosphatase 2C 39 [Jatropha curcas] |
| 6 |
Hb_089889_010 |
0.2641386961 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 [Jatropha curcas] |
| 7 |
Hb_004163_020 |
0.2655411162 |
- |
- |
- |
| 8 |
Hb_010288_050 |
0.2658325026 |
- |
- |
PREDICTED: uncharacterized protein LOC105642604 [Jatropha curcas] |
| 9 |
Hb_000975_140 |
0.2712351692 |
- |
- |
lactoylglutathione lyase family protein [Populus trichocarpa] |
| 10 |
Hb_001975_130 |
0.2722692077 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 6 isoform X1 [Populus euphratica] |
| 11 |
Hb_004116_120 |
0.2742168389 |
- |
- |
PREDICTED: protein EARLY FLOWERING 4 [Jatropha curcas] |
| 12 |
Hb_005375_050 |
0.277033685 |
- |
- |
inorganic pyrophosphatase, putative [Ricinus communis] |
| 13 |
Hb_044662_050 |
0.2780044564 |
- |
- |
hypothetical protein POPTR_0002s08440g [Populus trichocarpa] |
| 14 |
Hb_114292_010 |
0.2812407669 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000261_530 |
0.2839707214 |
- |
- |
PREDICTED: beta-glucosidase 18-like [Jatropha curcas] |
| 16 |
Hb_000165_270 |
0.2861540216 |
transcription factor |
TF Family: Tify |
JAZ10 [Hevea brasiliensis] |
| 17 |
Hb_005162_120 |
0.2868590187 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 18 |
Hb_002979_030 |
0.2873961847 |
- |
- |
PREDICTED: serine/threonine-protein kinase Aurora-1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_002007_040 |
0.2875239531 |
- |
- |
unnamed protein product [Coffea canephora] |
| 20 |
Hb_003399_100 |
0.28949286 |
- |
- |
PREDICTED: putative cyclin-A3-1 [Jatropha curcas] |