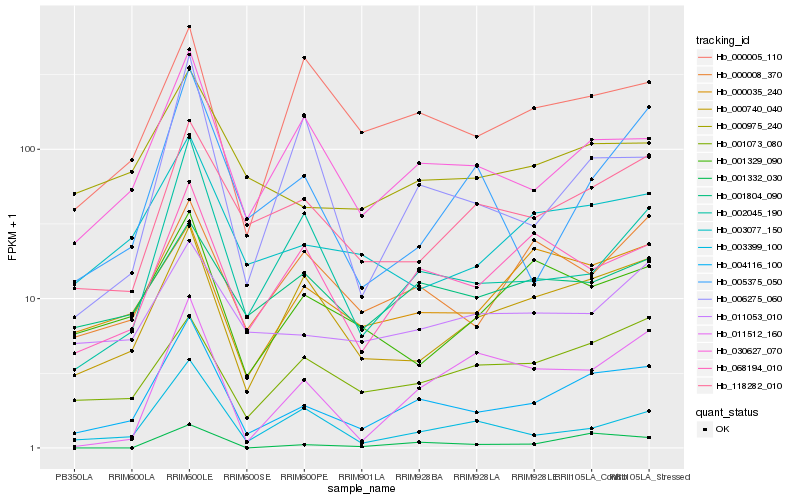
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004116_100 |
0.0 |
- |
- |
Isoamyl acetate-hydrolyzing esterase, putative [Ricinus communis] |
| 2 |
Hb_030627_070 |
0.1558841945 |
- |
- |
histone H4 [Zea mays] |
| 3 |
Hb_000740_040 |
0.1682510276 |
- |
- |
PREDICTED: uncharacterized protein LOC105126082 [Populus euphratica] |
| 4 |
Hb_003077_150 |
0.170407045 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |
| 5 |
Hb_118282_010 |
0.1761575881 |
- |
- |
PREDICTED: uncharacterized protein LOC105108358 [Populus euphratica] |
| 6 |
Hb_002045_190 |
0.1763579616 |
- |
- |
PREDICTED: histone H2A variant 1 [Jatropha curcas] |
| 7 |
Hb_068194_010 |
0.1793770461 |
- |
- |
hypothetical protein POPTR_0010s23740g [Populus trichocarpa] |
| 8 |
Hb_000975_240 |
0.1834941029 |
- |
- |
hypothetical protein JCGZ_22006 [Jatropha curcas] |
| 9 |
Hb_003399_100 |
0.1892285886 |
- |
- |
PREDICTED: putative cyclin-A3-1 [Jatropha curcas] |
| 10 |
Hb_001073_080 |
0.2002117829 |
- |
- |
PREDICTED: protein root UVB sensitive 2, chloroplastic [Jatropha curcas] |
| 11 |
Hb_011053_010 |
0.2019908429 |
- |
- |
PREDICTED: uncharacterized protein LOC105631146 [Jatropha curcas] |
| 12 |
Hb_001332_030 |
0.2024838951 |
- |
- |
hypothetical protein JCGZ_21111 [Jatropha curcas] |
| 13 |
Hb_001329_090 |
0.2024871406 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000005_110 |
0.203671864 |
- |
- |
histone H2B1 [Hevea brasiliensis] |
| 15 |
Hb_000008_370 |
0.2055304822 |
- |
- |
PREDICTED: uncharacterized protein LOC105628095 [Jatropha curcas] |
| 16 |
Hb_006275_060 |
0.2058319628 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 17 |
Hb_005375_050 |
0.2060504973 |
- |
- |
inorganic pyrophosphatase, putative [Ricinus communis] |
| 18 |
Hb_000035_240 |
0.2062383833 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_011512_160 |
0.2100758029 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001804_090 |
0.2108729041 |
- |
- |
PREDICTED: uracil phosphoribosyltransferase isoform X1 [Jatropha curcas] |