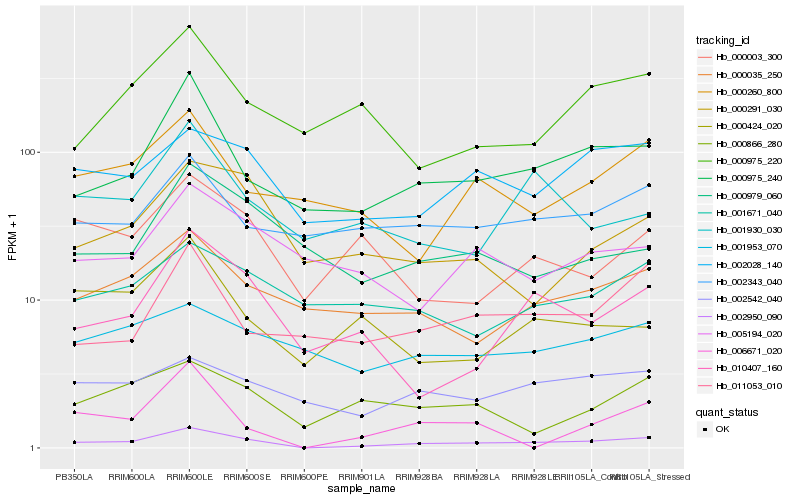
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002950_090 |
0.0 |
- |
- |
- |
| 2 |
Hb_000975_240 |
0.1787660045 |
- |
- |
hypothetical protein JCGZ_22006 [Jatropha curcas] |
| 3 |
Hb_002028_140 |
0.1974079459 |
transcription factor |
TF Family: Tify |
JAZ11 [Hevea brasiliensis] |
| 4 |
Hb_000035_250 |
0.2138777426 |
- |
- |
PREDICTED: uncharacterized protein LOC105637544 [Jatropha curcas] |
| 5 |
Hb_010407_160 |
0.2157636708 |
- |
- |
PREDICTED: chromatin modification-related protein MEAF6 isoform X1 [Jatropha curcas] |
| 6 |
Hb_011053_010 |
0.2166435398 |
- |
- |
PREDICTED: uncharacterized protein LOC105631146 [Jatropha curcas] |
| 7 |
Hb_002542_040 |
0.2184255767 |
- |
- |
hypothetical protein JCGZ_13884 [Jatropha curcas] |
| 8 |
Hb_002343_040 |
0.2201202925 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 2, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_000866_280 |
0.223520389 |
- |
- |
PREDICTED: seven transmembrane domain-containing tyrosine-protein kinase 1-like [Jatropha curcas] |
| 10 |
Hb_000260_800 |
0.2237728827 |
- |
- |
PREDICTED: probable NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 12 [Populus euphratica] |
| 11 |
Hb_000291_030 |
0.2308161123 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1-like [Eucalyptus grandis] |
| 12 |
Hb_000003_300 |
0.2309544007 |
- |
- |
PREDICTED: uncharacterized protein LOC105631182 [Jatropha curcas] |
| 13 |
Hb_000975_220 |
0.2313076896 |
- |
- |
Histone H2B [Medicago truncatula] |
| 14 |
Hb_001671_040 |
0.2315420919 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor RS31-like isoform X1 [Populus euphratica] |
| 15 |
Hb_001930_030 |
0.2323370394 |
- |
- |
PREDICTED: 30S ribosomal protein S10, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000979_060 |
0.2323826213 |
- |
- |
PREDICTED: protein PLANT CADMIUM RESISTANCE 2-like [Gossypium raimondii] |
| 17 |
Hb_000424_020 |
0.2333193933 |
- |
- |
Uncharacterized protein TCM_037386 [Theobroma cacao] |
| 18 |
Hb_005194_020 |
0.2334960714 |
- |
- |
PREDICTED: UPF0553 protein-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_006671_020 |
0.233798607 |
- |
- |
hypothetical protein CISIN_1g0407181mg, partial [Citrus sinensis] |
| 20 |
Hb_001953_070 |
0.2339788212 |
- |
- |
PREDICTED: RNA pseudouridine synthase 7 isoform X2 [Jatropha curcas] |