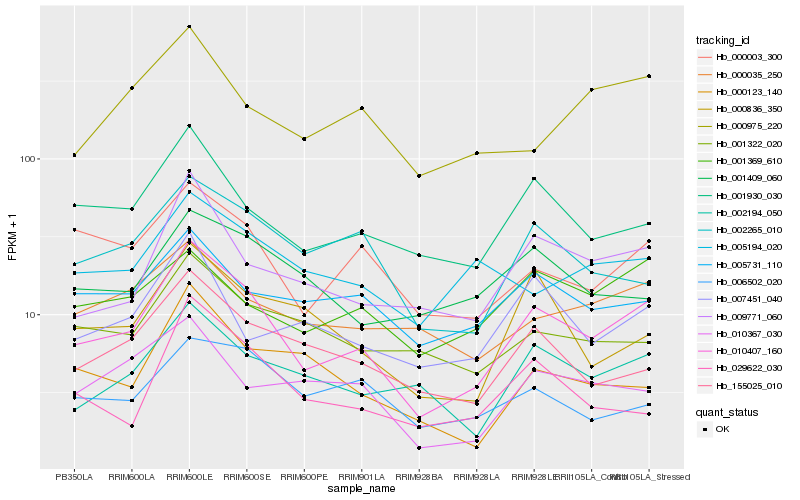
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010407_160 |
0.0 |
- |
- |
PREDICTED: chromatin modification-related protein MEAF6 isoform X1 [Jatropha curcas] |
| 2 |
Hb_009771_060 |
0.1485173501 |
- |
- |
PREDICTED: 30S ribosomal protein S31, chloroplastic [Populus euphratica] |
| 3 |
Hb_001930_030 |
0.1491344115 |
- |
- |
PREDICTED: 30S ribosomal protein S10, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000003_300 |
0.1565834876 |
- |
- |
PREDICTED: uncharacterized protein LOC105631182 [Jatropha curcas] |
| 5 |
Hb_002194_050 |
0.157352004 |
- |
- |
PREDICTED: UPF0187 protein At3g61320, chloroplastic [Jatropha curcas] |
| 6 |
Hb_155025_010 |
0.1579375774 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 7 |
Hb_002265_010 |
0.1688090252 |
- |
- |
ef-hand calcium binding protein, putative [Ricinus communis] |
| 8 |
Hb_005731_110 |
0.1718450998 |
- |
- |
PREDICTED: uncharacterized protein LOC105108367 isoform X1 [Populus euphratica] |
| 9 |
Hb_000035_250 |
0.1741170461 |
- |
- |
PREDICTED: uncharacterized protein LOC105637544 [Jatropha curcas] |
| 10 |
Hb_000836_350 |
0.1742602545 |
- |
- |
PREDICTED: uncharacterized protein LOC105642938 isoform X2 [Jatropha curcas] |
| 11 |
Hb_007451_040 |
0.1742943002 |
- |
- |
PREDICTED: uncharacterized protein LOC105643411 [Jatropha curcas] |
| 12 |
Hb_029622_030 |
0.176834674 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_001322_020 |
0.1777427408 |
- |
- |
PREDICTED: polycomb group protein VERNALIZATION 2-like isoform X2 [Populus euphratica] |
| 14 |
Hb_010367_030 |
0.1790347937 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000975_220 |
0.1799406337 |
- |
- |
Histone H2B [Medicago truncatula] |
| 16 |
Hb_006502_020 |
0.1801561471 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1-like [Jatropha curcas] |
| 17 |
Hb_000123_140 |
0.1803404631 |
- |
- |
PREDICTED: 21 kDa protein-like [Populus euphratica] |
| 18 |
Hb_001369_610 |
0.1818953266 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 2, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001409_060 |
0.1822964952 |
transcription factor |
TF Family: TRAF |
Regulatory protein NPR1, putative [Ricinus communis] |
| 20 |
Hb_005194_020 |
0.1840163753 |
- |
- |
PREDICTED: UPF0553 protein-like isoform X1 [Jatropha curcas] |