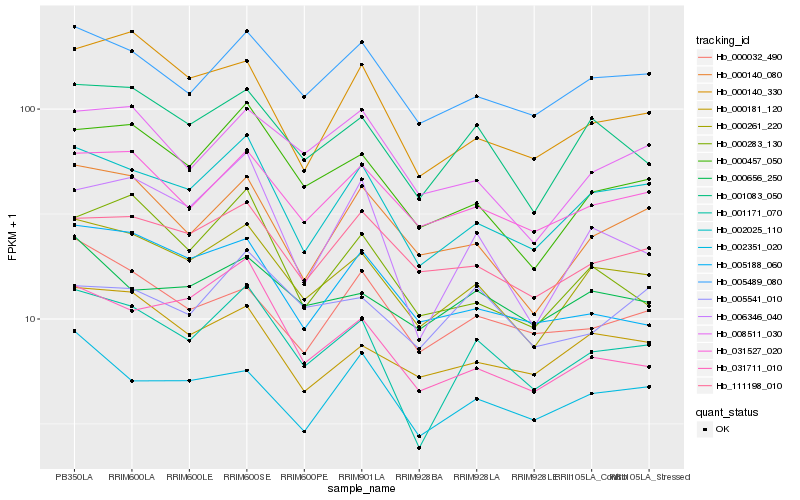
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001171_070 |
0.0 |
- |
- |
PREDICTED: 2-oxoisovalerate dehydrogenase subunit alpha 1, mitochondrial-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_000261_220 |
0.0787421999 |
- |
- |
PREDICTED: TBC1 domain family member 13 [Jatropha curcas] |
| 3 |
Hb_002025_110 |
0.0821833214 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: SWI/SNF complex component SNF12 homolog isoform X1 [Jatropha curcas] |
| 4 |
Hb_001083_050 |
0.0891880119 |
- |
- |
PREDICTED: V-type proton ATPase subunit d2 [Jatropha curcas] |
| 5 |
Hb_006346_040 |
0.0940073847 |
- |
- |
hypothetical protein JCGZ_20335 [Jatropha curcas] |
| 6 |
Hb_000457_050 |
0.0960804031 |
- |
- |
PREDICTED: tether containing UBX domain for GLUT4 [Jatropha curcas] |
| 7 |
Hb_005489_080 |
0.0985119446 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_031527_020 |
0.1064747996 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B isoform X1 [Jatropha curcas] |
| 9 |
Hb_000140_330 |
0.1132733147 |
- |
- |
PREDICTED: uncharacterized protein LOC105642909 [Jatropha curcas] |
| 10 |
Hb_002351_020 |
0.1151272948 |
- |
- |
PREDICTED: rRNA methyltransferase 3B, mitochondrial [Jatropha curcas] |
| 11 |
Hb_031711_010 |
0.1159791802 |
- |
- |
PREDICTED: CDPK-related kinase 4-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000032_490 |
0.1161446237 |
- |
- |
PREDICTED: uncharacterized protein At4g10930-like [Jatropha curcas] |
| 13 |
Hb_000140_080 |
0.1170017269 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 3-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_008511_030 |
0.1181520023 |
- |
- |
guanine nucleotide-binding protein beta, putative [Ricinus communis] |
| 15 |
Hb_000181_120 |
0.1187140499 |
- |
- |
PREDICTED: PRA1 family protein H isoform X1 [Jatropha curcas] |
| 16 |
Hb_000656_250 |
0.1197415329 |
- |
- |
hypothetical protein JCGZ_24680 [Jatropha curcas] |
| 17 |
Hb_111198_010 |
0.1208899506 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 37-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_005188_060 |
0.1209234547 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 19 |
Hb_005541_010 |
0.1211708747 |
- |
- |
PREDICTED: uncharacterized protein LOC105629250 [Jatropha curcas] |
| 20 |
Hb_000283_130 |
0.1214727842 |
- |
- |
PREDICTED: PRA1 family protein E-like [Populus euphratica] |