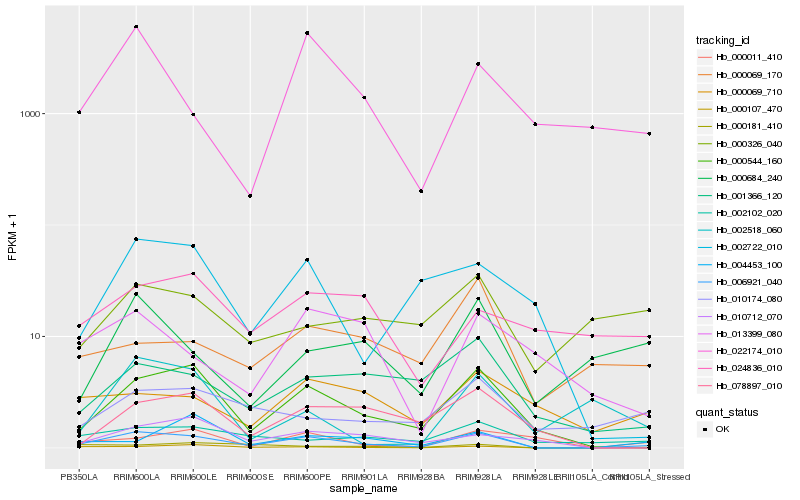
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006921_040 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g25630 [Jatropha curcas] |
| 2 |
Hb_000544_160 |
0.2145527261 |
- |
- |
PREDICTED: phosphomethylethanolamine N-methyltransferase-like [Nelumbo nucifera] |
| 3 |
Hb_000107_470 |
0.2568563225 |
- |
- |
cellulose synthase, putative [Ricinus communis] |
| 4 |
Hb_001366_120 |
0.2818255795 |
- |
- |
unknown [Zea mays] |
| 5 |
Hb_002102_020 |
0.2950015899 |
- |
- |
PREDICTED: lysine-specific histone demethylase 1 homolog 1 [Jatropha curcas] |
| 6 |
Hb_010174_080 |
0.2961481561 |
- |
- |
- |
| 7 |
Hb_000181_410 |
0.2988186725 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_022174_010 |
0.3002909191 |
- |
- |
elicitor-responsive protein [Hevea brasiliensis] |
| 9 |
Hb_002722_010 |
0.3008927897 |
- |
- |
Disease resistance response protein, putative [Ricinus communis] |
| 10 |
Hb_078897_010 |
0.3013329838 |
- |
- |
hypothetical protein POPTR_0012s044902g, partial [Populus trichocarpa] |
| 11 |
Hb_000011_410 |
0.3017582571 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_010712_070 |
0.3020458672 |
- |
- |
Galactoside 2-alpha-L-fucosyltransferase [Aegilops tauschii] |
| 13 |
Hb_000684_240 |
0.306250245 |
- |
- |
hypothetical protein RCOM_0819620 [Ricinus communis] |
| 14 |
Hb_000069_710 |
0.3099363424 |
- |
- |
PREDICTED: uncharacterized protein LOC105642890 [Jatropha curcas] |
| 15 |
Hb_004453_100 |
0.3132878305 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_013399_080 |
0.3155877732 |
- |
- |
PREDICTED: protein Brevis radix-like 2 [Jatropha curcas] |
| 17 |
Hb_000069_170 |
0.319905686 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 18 [Jatropha curcas] |
| 18 |
Hb_024836_010 |
0.3210719785 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 19 |
Hb_000326_040 |
0.3247855378 |
- |
- |
hypothetical protein PHAVU_006G187900g [Phaseolus vulgaris] |
| 20 |
Hb_002518_060 |
0.3281743756 |
- |
- |
PREDICTED: uncharacterized protein LOC105631129 [Jatropha curcas] |