| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
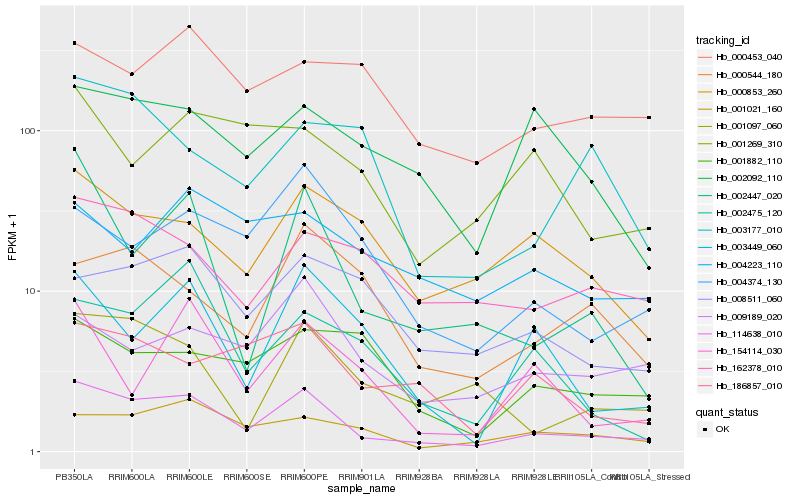
Hb_114638_010 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62930, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_001021_160 |
0.1959519157 |
- |
- |
PREDICTED: homeobox protein LUMINIDEPENDENS [Jatropha curcas] |
| 3 |
Hb_001097_060 |
0.1967129387 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 8 [Jatropha curcas] |
| 4 |
Hb_002447_020 |
0.201744148 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000853_260 |
0.2032236024 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 7 [Jatropha curcas] |
| 6 |
Hb_162378_010 |
0.2035388361 |
- |
- |
hypothetical protein JCGZ_20480 [Jatropha curcas] |
| 7 |
Hb_009189_020 |
0.2035534425 |
- |
- |
PREDICTED: CASP-like protein 4B4 [Jatropha curcas] |
| 8 |
Hb_186857_010 |
0.2039977239 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 9 |
Hb_003449_060 |
0.2054508226 |
- |
- |
PREDICTED: L-ascorbate oxidase homolog [Jatropha curcas] |
| 10 |
Hb_004374_130 |
0.2055764312 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA1B-like [Jatropha curcas] |
| 11 |
Hb_008511_060 |
0.2113239268 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At4g34500 [Jatropha curcas] |
| 12 |
Hb_002092_110 |
0.2128050836 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 1 [Jatropha curcas] |
| 13 |
Hb_002475_120 |
0.2162227645 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 14 |
Hb_001269_310 |
0.2216600679 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 6 [Jatropha curcas] |
| 15 |
Hb_154114_030 |
0.2223074701 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 16 |
Hb_000544_180 |
0.2234890222 |
- |
- |
PREDICTED: probable auxin efflux carrier component 1c isoform X1 [Jatropha curcas] |
| 17 |
Hb_000453_040 |
0.2257685276 |
- |
- |
Triosephosphate isomerase, cytosolic [Gossypium arboreum] |
| 18 |
Hb_004223_110 |
0.2277859751 |
- |
- |
PREDICTED: putative HVA22-like protein g [Populus euphratica] |
| 19 |
Hb_001882_110 |
0.2282813353 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003177_010 |
0.2300215916 |
- |
- |
PREDICTED: putative HVA22-like protein g [Populus euphratica] |